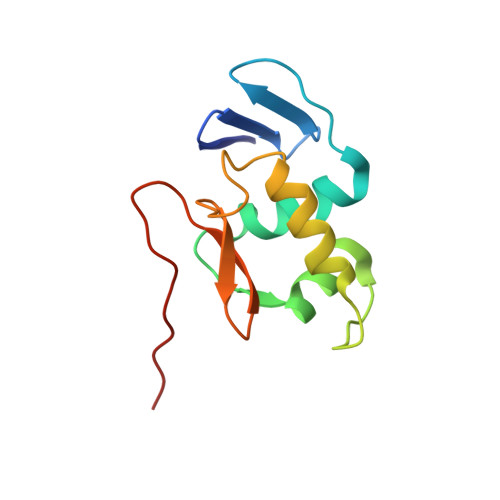
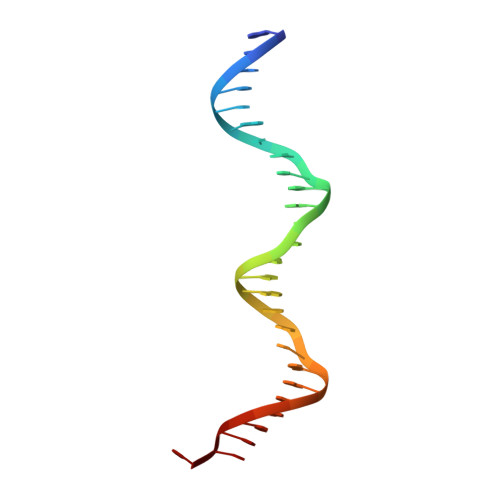
Structural basis for promoter DNA recognition by the response regulator OmpR.
Sadotra, S., Lou, Y.C., Tang, H.C., Chiu, Y.C., Hsu, C.H., Chen, C.(2020) J Struct Biol 213: 107638-107638
- PubMed: 33152421
- DOI: https://doi.org/10.1016/j.jsb.2020.107638
- Primary Citation of Related Structures:
6LXL, 6LXM, 6LXN - PubMed Abstract:
OmpR, a response regulator of the EnvZ/OmpR two-component system (TCS), controls the reciprocal regulation of two porin proteins, OmpF and OmpC, in bacteria. During signal transduction, OmpR (OmpR-FL) undergoes phosphorylation at its conserved Asp residue in the N-terminal receiver domain (OmpRn) and recognizes the promoter DNA from its C-terminal DNA-binding domain (OmpRc) to elicit an adaptive response. Apart from that, OmpR regulates many genes in Escherichia coli and is important for virulence in several pathogens. However, the molecular mechanism of the regulation and the structural basis of OmpR-DNA binding is still not fully clear. In this study, we presented the crystal structure of OmpRc in complex with the F1 region of the ompF promoter DNA from E. coli. Our structural analysis suggested that OmpRc binds to its cognate DNA as a homodimer, only in a head-to-tail orientation. Also, the OmpRc apo-form showed a unique domain-swapped crystal structure under different crystallization conditions. Biophysical experimental data, such as NMR, fluorescent polarization and thermal stability, showed that inactive OmpR-FL (unphosphorylated) could bind to promoter DNA with a weaker binding affinity as compared with active OmpR-FL (phosphorylated) or OmpRc, and also confirmed that phosphorylation may only enhance DNA binding. Furthermore, the dimerization interfaces in the OmpRc-DNA complex structure identified in this study provide an opportunity to understand the regulatory role of OmpR and explore the potential for this "druggable" target.
- Institute of Biomedical Sciences, Academia Sinica, Taipei 115, Taiwan; Chemical Biology and Molecular Biophysics, Taiwan International Graduate Program, Academia Sinica, Taipei 115, Taiwan; Institute of Bioinformatics and Structural Biology, National Tsing Hua University, Hsinchu 300, Taiwan. Electronic address: sushant@gate.sinica.edu.tw.
Organizational Affiliation: