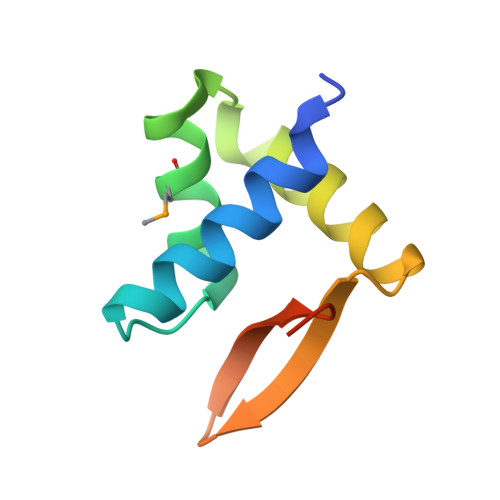
The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Stielow, B., Zhou, Y., Cao, Y., Simon, C., Pogoda, H.M., Jiang, J., Ren, Y., Phanor, S.K., Rohner, I., Nist, A., Stiewe, T., Hammerschmidt, M., Shi, Y., Bulyk, M.L., Wang, Z., Liefke, R.(2021) Sci Adv 7
- PubMed: 33980486
- DOI: https://doi.org/10.1126/sciadv.abf2229
- Primary Citation of Related Structures:
6LUI, 6LUJ, 6LUK - PubMed Abstract:
CpG islands (CGIs) are key regulatory DNA elements at most promoters, but how they influence the chromatin status and transcription remains elusive. Here, we identify and characterize SAMD1 (SAM domain-containing protein 1) as an unmethylated CGI-binding protein. SAMD1 has an atypical winged-helix domain that directly recognizes unmethylated CpG-containing DNA via simultaneous interactions with both the major and the minor groove. The SAM domain interacts with L3MBTL3, but it can also homopolymerize into a closed pentameric ring. At a genome-wide level, SAMD1 localizes to H3K4me3-decorated CGIs, where it acts as a repressor. SAMD1 tethers L3MBTL3 to chromatin and interacts with the KDM1A histone demethylase complex to modulate H3K4me2 and H3K4me3 levels at CGIs, thereby providing a mechanism for SAMD1-mediated transcriptional repression. The absence of SAMD1 impairs ES cell differentiation processes, leading to misregulation of key biological pathways. Together, our work establishes SAMD1 as a newly identified chromatin regulator acting at unmethylated CGIs.
- Institute of Molecular Biology and Tumor Research (IMT), Philipps University of Marburg, 35043 Marburg, Germany.
Organizational Affiliation: