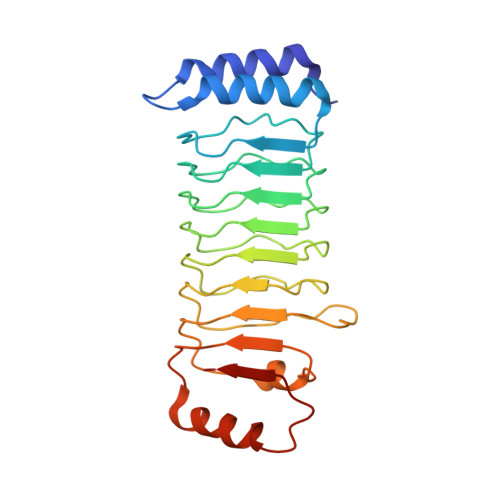
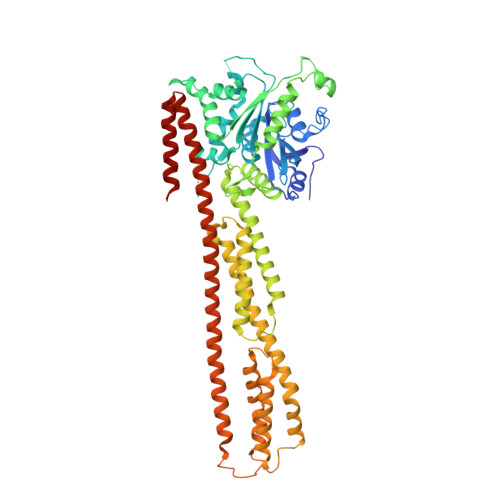
Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Ye, Y., Xiong, Y., Huang, H.(2020) Commun Biol 3: 752-752
- PubMed: 33303953
- DOI: https://doi.org/10.1038/s42003-020-01492-1
- Primary Citation of Related Structures:
6LOJ, 6LOL - PubMed Abstract:
IpaH enzymes are bacterial E3 ligases targeting host proteins for ubiquitylation. Two autoinhibition modes of IpaH enzymes have been proposed based on the relative positioning of the Leucine-rich repeat domain (LRR) with respect to the NEL domain. In mode 1, substrate-binding competitively displaces the interactions between theLRR and NEL to relieve autoinhibition. However, the molecular basis for mode 2 is unclear. Here, we present the crystal structures of Shigella IpaH9.8 and the LRR of IpaH9.8 in complex with the substrate of human guanylate-binding protein 1 (hGBP1). A hydrophobic cluster in the C-terminus of IpaH9.8 LRR forms a hydrophobic pocket involved in binding the NEL domain, and the binding is important for IpaH9.8 autoinhibition. Substrate-binding destabilizes the hydrophobic cluster by inducing conformational changes of IpaH9.8 LRR . Arg166 and Phe187 in IpaH9.8 LRR function as sensors for substrate-binding. Collectively, our findings provide insights into the molecular mechanisms for the actication of IpaH9.8 in autoinhibition mode 2.
- State Key Laboratory of Chemical Oncogenomics, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, 518055, Shenzhen, China. yeyx@pkusz.edu.cn.
Organizational Affiliation: