Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Li, J., Yang, J.G., Mu, S., Shang, N., Liu, C., Zhu, Y., Cai, Y., Liu, P., Lin, J., Liu, W.D., Sun, Y.X., Ma, Y.(2020) ACS Catal
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glycosyltransferase | 454 | Siraitia grosvenorii | Mutation(s): 0 Gene Names: UDPG1 EC: 2.4.1 (PDB Primary Data), 2.4.1.350 (UniProt) | 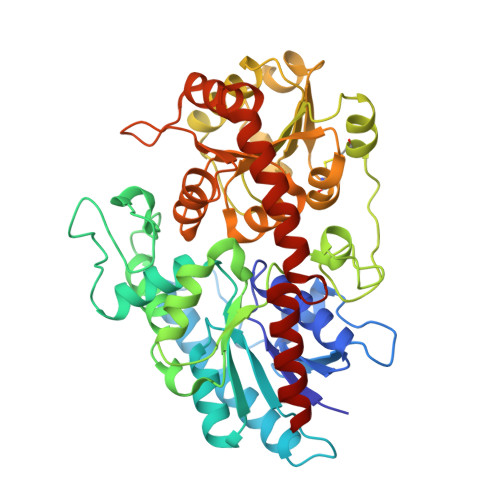 | |
UniProt | |||||
Find proteins for K7NBW3 (Siraitia grosvenorii) Explore K7NBW3 Go to UniProtKB: K7NBW3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K7NBW3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | B [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 50.832 | α = 90 |
| b = 68.877 | β = 90 |
| c = 143.908 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (China) | China | -- |