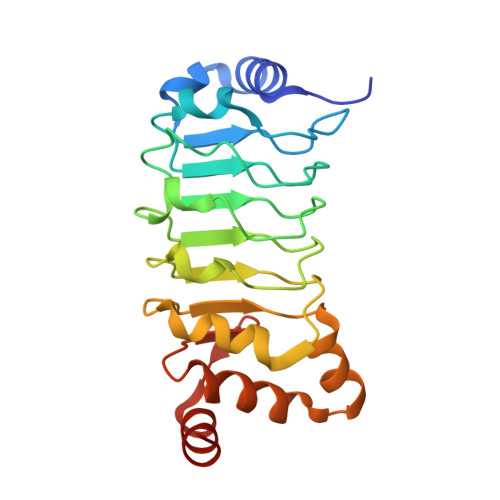
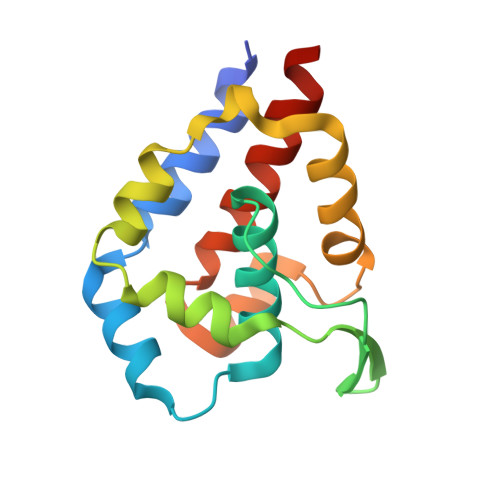
The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
Toda, A., Nishikawa, Y., Tanaka, H., Yagi, T., Kurisu, G.(2020) J Biological Chem 295: 3982-3989
- PubMed: 32014992
- DOI: https://doi.org/10.1074/jbc.RA119.011541
- Primary Citation of Related Structures:
6L4P - PubMed Abstract:
Axonemal dynein is a microtubule-based molecular motor that drives ciliary/flagellar beating in eukaryotes. In axonemal dynein, the outer-arm dynein (OAD) complex, which comprises three heavy chains (α, β, and γ), produces the main driving force for ciliary/flagellar motility. It has recently been shown that axonemal dynein light chain-1 (LC1) binds to the microtubule-binding domain (MTBD) of OADγ, leading to a decrease in its microtubule-binding affinity. However, it remains unclear how LC1 interacts with the MTBD and controls the microtubule-binding affinity of OADγ. Here, we have used X-ray crystallography and pulldown assays to examine the interaction between LC1 and the MTBD, identifying two important sites of interaction in the MTBD. Solving the LC1-MTBD complex from Chlamydomonas reinhardtii at 1.7 Å resolution, we observed that one site is located in the H5 helix and that the other is located in the flap region that is unique to some axonemal dynein MTBDs. Mutational analysis of key residues in these sites indicated that the H5 helix is the main LC1-binding site. We modeled the ternary structure of the LC1-MTBD complex bound to microtubules based on the known dynein-microtubule complex. This enabled us to propose a structural basis for both formations of the ternary LC1-MTBD-microtubule complex and LC1-mediated tuning of MTBD binding to the microtubule, suggesting a molecular model for how axonemal dynein senses the curvature of the axoneme and tunes ciliary/flagellar beating.
- Department of Biological Sciences, Osaka University, Toyonaka, Osaka 560-0043, Japan.
Organizational Affiliation: