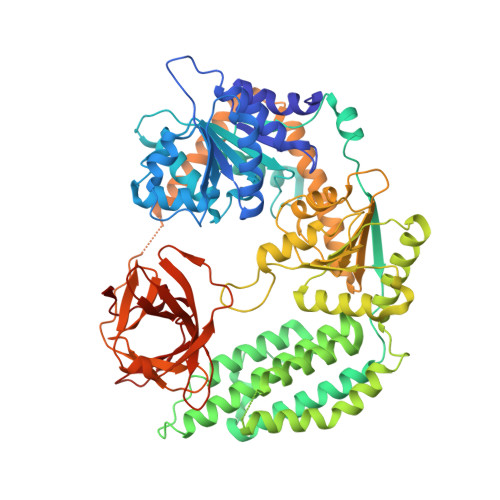
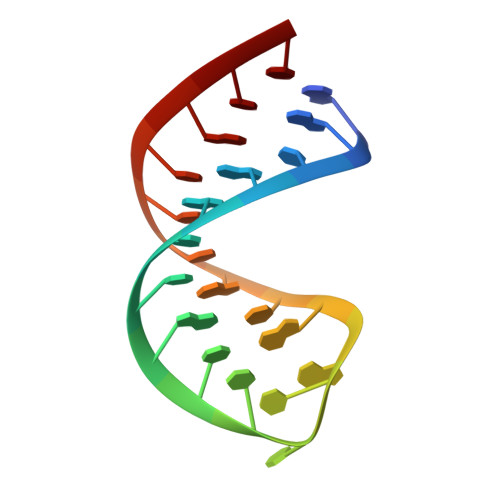
Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Kim, K.H., Hwang, J., Kim, J.H., Son, K.P., Jang, Y., Kim, M., Kang, S.J., Lee, J.O., Kang, J.Y., Choi, B.S.(2020) RNA Biol 17: 325-334
- PubMed: 31852354
- DOI: https://doi.org/10.1080/15476286.2019.1700034
- Primary Citation of Related Structures:
6KYV - PubMed Abstract:
Retinoic acid-inducible gene I (RIG-I) is responsible for innate immunity via the recognition of short double-stranded RNAs in the cytosol. With the clue that G-U wobble base pairs in the influenza A virus's RNA promoter region are responsible for RIG-I activation, we determined the complex structure of RIG-I ΔCARD and a short hairpin RNA with G-U wobble base pairs by X-ray crystallography. Interestingly, the overall helical backbone trace was not affected by the presence of the wobble base pairs; however, the base pair inclination and helical axis angle changed upon RIG-I binding. NMR spectroscopy revealed that RIG-I binding renders the flexible base pair of the influenza A virus's RNA promoter region between the two G-U wobble base pairs even more flexible. Binding to RNA with wobble base pairs resulted in a more flexible RIG-I complex. This flexible complex formation correlates with the entropy-favoured binding of RIG-I and RNA, which results in tighter binding affinity and RIG-I activation. This study suggests that the structure and dynamics of RIG-I are tailored to the binding of specific RNA sequences with different flexibility.
- Department of Chemistry, KAIST, Daejeon, Republic of Korea.
Organizational Affiliation: