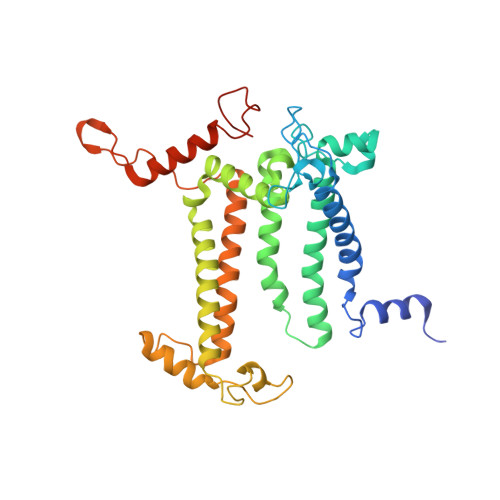
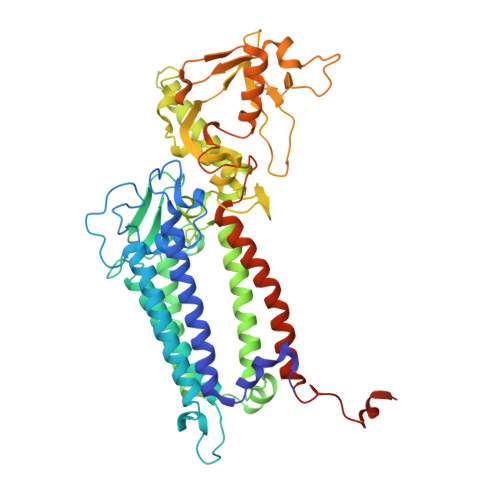
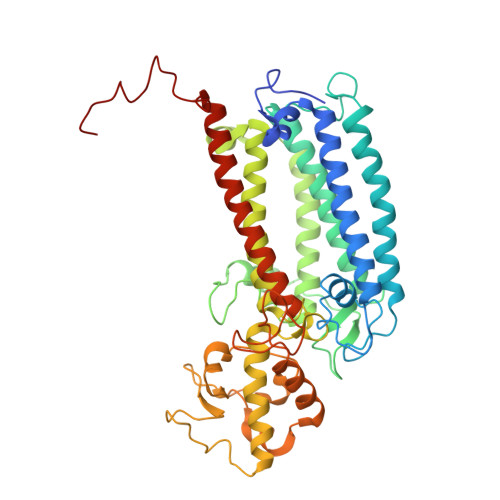
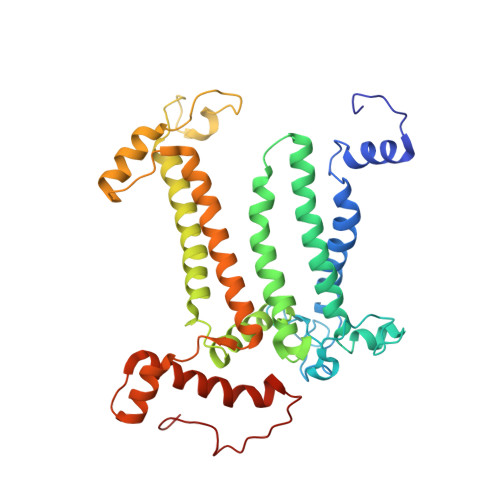
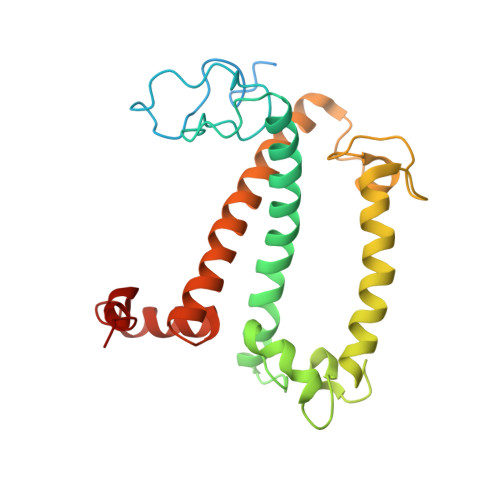
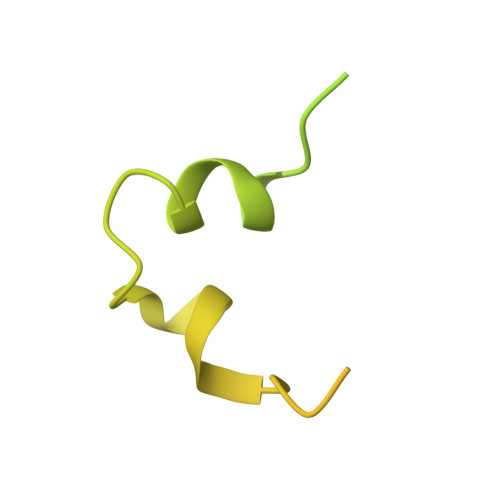
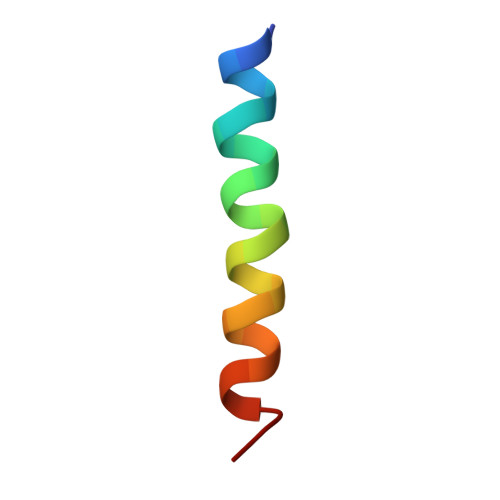Structural insight into light harvesting for photosystem II in green algae.
Sheng, X., Watanabe, A., Li, A., Kim, E., Song, C., Murata, K., Song, D., Minagawa, J., Liu, Z.(2019) Nat Plants 5: 1320-1330
- PubMed: 31768031
- DOI: https://doi.org/10.1038/s41477-019-0543-4
- Primary Citation of Related Structures:
6KAC, 6KAD - PubMed Abstract:
Green algae and plants rely on light-harvesting complex II (LHCII) to collect photon energy for oxygenic photosynthesis. In Chlamydomonas reinhardtii, LHCII molecules associate with photosystem II (PSII) to form various supercomplexes, including the C 2 S 2 M 2 L 2 type, which is the largest PSII-LHCII supercomplex in algae and plants that is presently known. Here, we report high-resolution cryo-electron microscopy (cryo-EM) maps and structural models of the C 2 S 2 M 2 L 2 and C 2 S 2 supercomplexes from C. reinhardtii. The C 2 S 2 supercomplex contains an LhcbM1-LhcbM2/7-LhcbM3 heterotrimer in the strongly associated LHCII, and the LhcbM1 subunit assembles with CP43 through two interfacial galactolipid molecules. The loosely and moderately associated LHCII trimers interact closely with the minor antenna complex CP29 to form an intricate subcomplex bound to CP47 in the C 2 S 2 M 2 L 2 supercomplex. A notable direct pathway is established for energy transfer from the loosely associated LHCII to the PSII reaction centre, as well as several indirect routes. Structure-based computational analysis on the excitation energy transfer within the two supercomplexes provides detailed mechanistic insights into the light-harvesting process in green algae.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: