Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Masuno, H., Kazui, Y., Tanatani, A., Fujii, S., Kawachi, E., Ikura, T., Ito, N., Yamamoto, K., Kagechika, H.(2019) Bioorg Med Chem 27: 3674-3681
- PubMed: 31300316
- DOI: https://doi.org/10.1016/j.bmc.2019.07.003
- Primary Citation of Related Structures:
6K5O - PubMed Abstract:
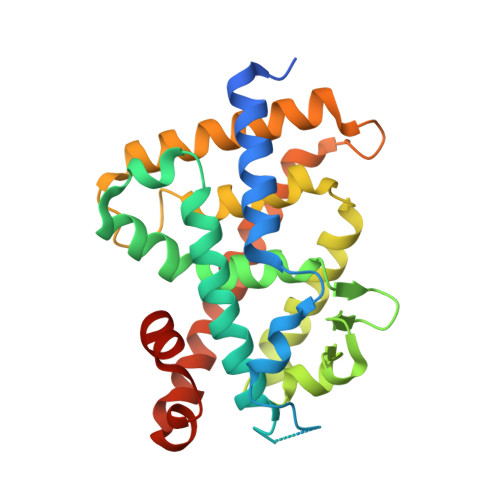
Lithocholic acid (2) was identified as the second endogenous ligand of vitamin D receptor (VDR), though its binding affinity to VDR and its vitamin D activity are very weak compared to those of the active metabolite of vitamin D 3 , 1α,25-dihydroxyvitamin D 3 (1). 3-Acylated lithocholic acids were reported to be slightly more potent than lithocholic acid (2) as VDR agonists. Here, aiming to develop more potent lithocholic acid derivatives, we synthesized several derivatives bearing a 3-sulfonate/carbonate or 3-amino/amide substituent, and examined their differentiation-inducing activity toward human promyelocytic leukemia HL-60 cells. Introduction of a nitrogen atom at the 3-position of lithocholic acid (2) decreased the activity, but compound 6 bearing a 3-methylsulfonate group showed more potent activity than lithocholic acid (2) or its acylated derivatives. The binding of 6 to VDR was confirmed by competitive binding assay and X-ray crystallographic analysis of the complex of VDR ligand-binding domain (LBD) with 6.
- Institute of Biomaterials and Bioengineering, Tokyo Medical and Dental University, 2-3-10 Kanda-Surugadai, Chiyoda-ku, Tokyo 101-0062, Japan.
Organizational Affiliation: