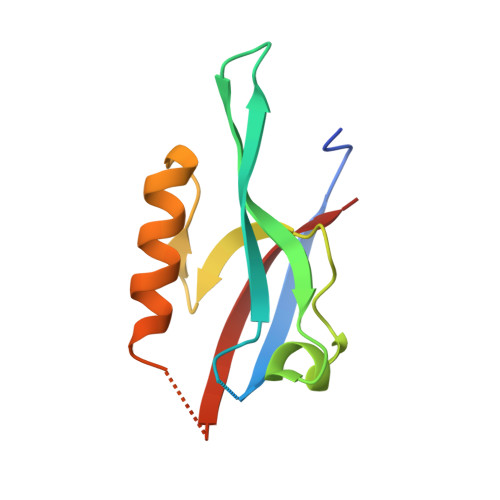
Par complex cluster formation mediated by phase separation.
Liu, Z., Yang, Y., Gu, A., Xu, J., Mao, Y., Lu, H., Hu, W., Lei, Q.Y., Li, Z., Zhang, M., Cai, Y., Wen, W.(2020) Nat Commun 11: 2266-2266
- PubMed: 32385244
- DOI: https://doi.org/10.1038/s41467-020-16135-6
- Primary Citation of Related Structures:
6JUE - PubMed Abstract:
The evolutionarily conserved Par3/Par6/aPKC complex regulates the polarity establishment of diverse cell types and distinct polarity-driven functions. However, how the Par complex is concentrated beneath the membrane to initiate cell polarization remains unclear. Here we show that the Par complex exhibits cell cycle-dependent condensation in Drosophila neuroblasts, driven by liquid-liquid phase separation. The open conformation of Par3 undergoes autonomous phase separation likely due to its NTD-mediated oligomerization. Par6, via C-terminal tail binding to Par3 PDZ3, can be enriched to Par3 condensates and in return dramatically promote Par3 phase separation. aPKC can also be concentrated to the Par3N/Par6 condensates as a client. Interestingly, activated aPKC can disperse the Par3/Par6 condensates via phosphorylation of Par3. Perturbations of Par3/Par6 phase separation impair the establishment of apical-basal polarity during neuroblast asymmetric divisions and lead to defective lineage development. We propose that phase separation may be a common mechanism for localized cortical condensation of cell polarity complexes.
- Department of Neurosurgery, Huashan Hospital, the Shanghai Key Laboratory of Medical Epigenetics, the International Co-laboratory of Medical Epigenetics and Metabolism, Ministry of Science and Technology, Institutes of Biomedical Sciences, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, School of Basic Medical Sciences, Fudan University, Shanghai, 200032, China.
Organizational Affiliation: