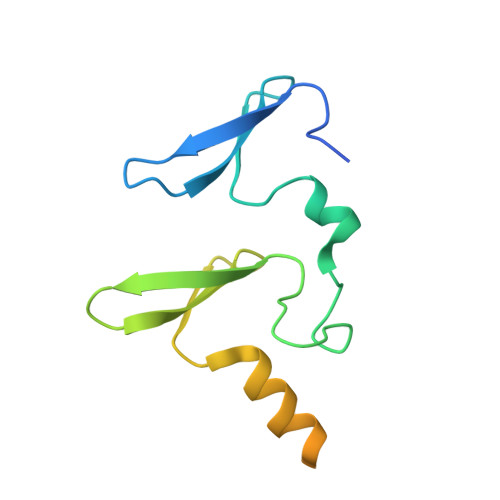
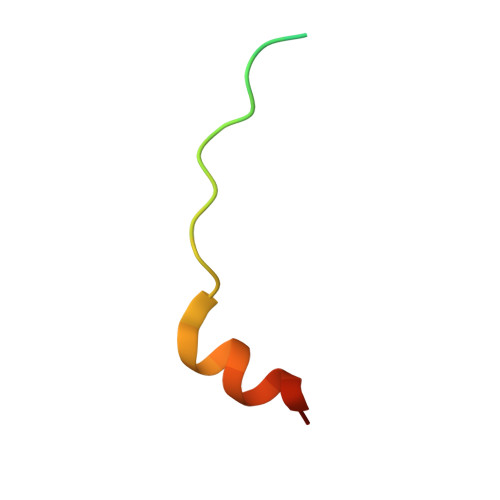
Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Lin, Z., Yang, Z., Xie, R., Ji, Z., Guan, K., Zhang, M.(2019) Elife 8
- PubMed: 31486770
- DOI: https://doi.org/10.7554/eLife.49439
- Primary Citation of Related Structures:
6J68, 6JJW, 6JJX, 6JJY, 6JJZ, 6JK0, 6JK1 - PubMed Abstract:
WW domain tandem-containing proteins such as KIBRA, YAP, and MAGI play critical roles in cell growth and polarity via binding to and positioning target proteins in specific subcellular regions. An immense disparity exists between promiscuity of WW domain-mediated target bindings and specific roles of WW domain proteins in cell growth regulation. Here, we discovered that WW domain tandems of KIBRA and MAGI, but not YAP, bind to specific target proteins with extremely high affinity and exquisite sequence specificity. Via systematic structural biology and biochemistry approaches, we decoded the target binding rules of WW domain tandems from cell growth regulatory proteins and uncovered a list of previously unknown WW tandem binding proteins including β-Dystroglycan, JCAD, and PTPN21. The WW tandem-mediated target recognition mechanisms elucidated here can guide functional studies of WW domain proteins in cell growth and polarity as well as in other cellular processes including neuronal synaptic signaling.
- Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Hong Kong, China.
Organizational Affiliation: