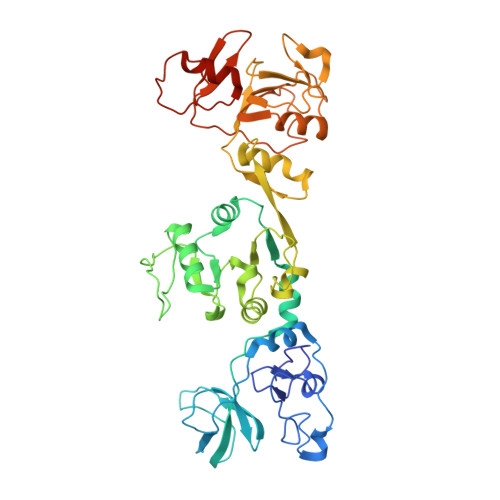
Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Tang, C., Deng, Z., Li, X., Yang, M., Tian, Z., Chen, Z., Wang, G., Wu, W., Feng, W.H., Zhang, G., Chen, Z.(2020) Viruses 12
- PubMed: 32075207
- DOI: https://doi.org/10.3390/v12020215
- Primary Citation of Related Structures:
6JDR, 6JDS, 6JDU - PubMed Abstract:
Porcine reproductive and respiratory syndrome virus (PRRSV) is prevalent throughout the world and has caused great economic losses to the swine industry. Nonstructural protein 10 (nsp10) is a superfamily 1 helicase participating in multiple processes of virus replication and one of the three most conserved proteins in nidoviruses. Here we report three high resolution crystal structures of highly pathogenic PRRSV nsp10. PRRSV nsp10 has multiple domains, including an N-terminal zinc-binding domain (ZBD), a β-barrel domain, a helicase core with two RecA-like domains, and a C-terminal domain (CTD). The CTD adopts a novel fold and is required for the overall structure and enzymatic activities. Although each domain except the CTD aligns well with its homologs, PRRSV nsp10 adopts an unexpected extended overall structure in crystals and solution. Moreover, structural and functional analyses of PRRSV nsp10 versus its closest homolog, equine arteritis virus nsp10, suggest that DNA binding might induce a profound conformational change of PRRSV nsp10 to exert functions, thus shedding light on the mechanisms of activity regulation of this helicase.
- State Key Laboratory of Agrobiotechnology and Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Organizational Affiliation: