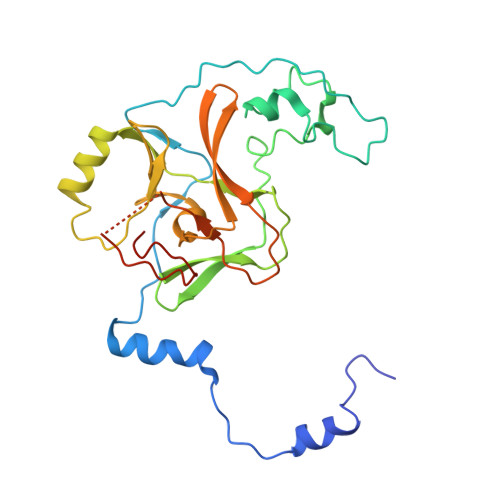
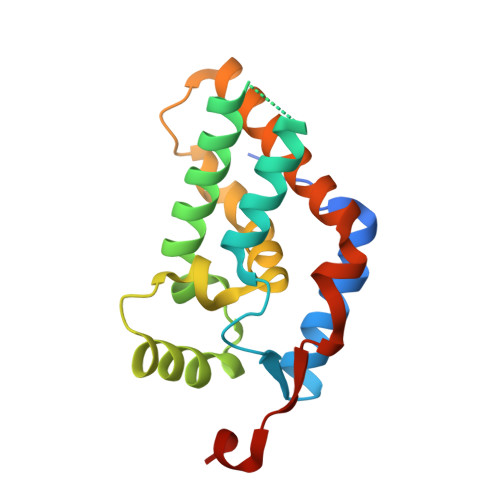
Structural Insights into Stimulation of Ash1L's H3K36 Methyltransferase Activity through Mrg15 Binding.
Hou, P., Huang, C., Liu, C.P., Yang, N., Yu, T., Yin, Y., Zhu, B., Xu, R.M.(2019) Structure 27: 837
- PubMed: 30827843
- DOI: https://doi.org/10.1016/j.str.2019.01.015
- Primary Citation of Related Structures:
6INE - PubMed Abstract:
The evolutionarily conserved Trithorax group protein Ash1 is a SET domain histone methyltransferase that mono- and dimethylates lysine 36 of histone H3 (H3K36). Ash1 forms a complex with Mrg15 and Nurf55, and the binding of Mrg15 greatly stimulates the catalytic activity of Ash1, yet the underlying molecular mechanisms remain unknown. Here we report the crystal structure of the tandem Mrg15-interacting and SET domains of human Ash1L in complex with Mrg15. Ash1L interacts with Mrg15 principally via a segment located N-terminal to the catalytic SET domain. Surprisingly, an autoinhibitory loop in the post-SET region of Ash1L is destabilized on Mrg15 binding despite no direct contact. Dynamics of the autoinhibitory loop can be attributed to subtle structural changes of the S-adenosylmethionine (SAM) binding pocket induced by Mrg15 binding, implicating a mechanism of conformational coupling between SAM and substrate binding sites. The findings broaden the understanding of regulation of H3K36 methyltransferases.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 10049, China.
Organizational Affiliation: