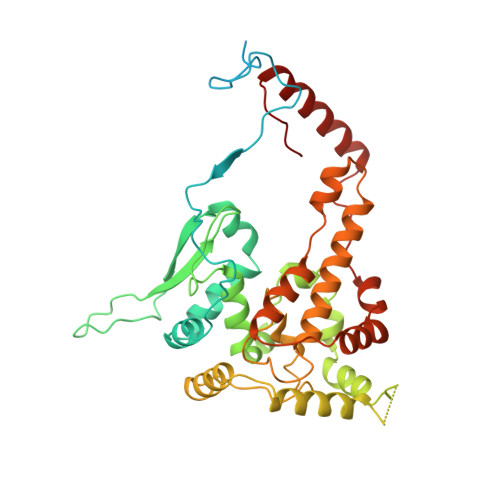
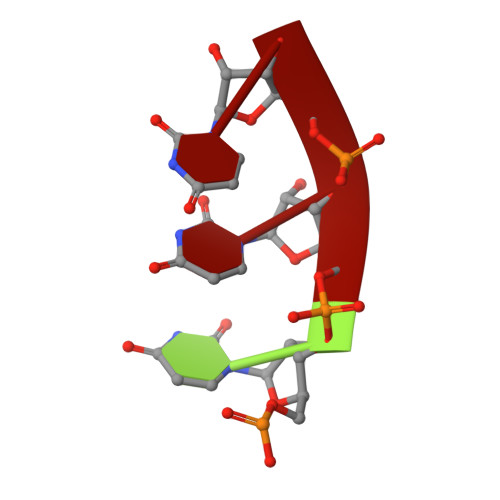
High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms.
Arragain, B., Reguera, J., Desfosses, A., Gutsche, I., Schoehn, G., Malet, H.(2019) Elife 8
- PubMed: 30638449
- DOI: https://doi.org/10.7554/eLife.43075
- Primary Citation of Related Structures:
6I2N - PubMed Abstract:
Negative-strand RNA viruses condense their genome into helical nucleocapsids that constitute essential templates for viral replication and transcription. The intrinsic flexibility of nucleocapsids usually prevents their full-length structural characterisation at high resolution. Here, we describe purification of full-length recombinant metastable helical nucleocapsid of Hantaan virus ( Hantaviridae family, Bunyavirales order) and determine its structure at 3.3 Å resolution by cryo-electron microscopy. The structure reveals the mechanisms of helical multimerisation via sub-domain exchanges between protomers and highlights nucleotide positions in a continuous positively charged groove compatible with viral genome binding. It uncovers key sites for future structure-based design of antivirals that are currently lacking to counteract life-threatening hantavirus infections. The structure also suggests a model of nucleoprotein-polymerase interaction that would enable replication and transcription solely upon local disruption of the nucleocapsid.
- Electron Microscopy and Methods Group, Université Grenoble Alpes, CNRS, CEA, Institute for Structural Biology, Grenoble, France.
Organizational Affiliation: