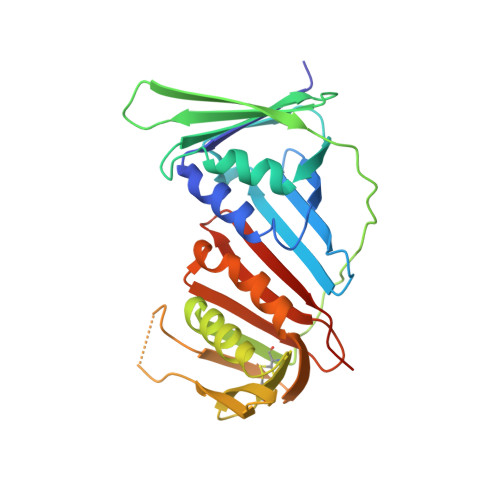
The p12 subunit of human polymerase delta uses an atypical PIP box for molecular recognition of proliferating cell nuclear antigen (PCNA).
Gonzalez-Magana, A., Ibanez de Opakua, A., Romano-Moreno, M., Murciano-Calles, J., Merino, N., Luque, I., Rojas, A.L., Onesti, S., Blanco, F.J., De Biasio, A.(2019) J Biological Chem 294: 3947-3956
- PubMed: 30655288
- DOI: https://doi.org/10.1074/jbc.RA118.006391
- Primary Citation of Related Structures:
6HVO - PubMed Abstract:
Human DNA polymerase δ is essential for DNA replication and acts in conjunction with the processivity factor proliferating cell nuclear antigen (PCNA). In addition to its catalytic subunit (p125), pol δ comprises three regulatory subunits (p50, p68, and p12). PCNA interacts with all of these subunits, but only the interaction with p68 has been structurally characterized. Here, we report solution NMR-, isothermal calorimetry-, and X-ray crystallography-based analyses of the p12-PCNA interaction, which takes part in the modulation of the rate and fidelity of DNA synthesis by pol δ. We show that p12 binds with micromolar affinity to the classical PIP-binding pocket of PCNA via a highly atypical PIP box located at the p12 N terminus. Unlike the canonical PIP box of p68, the PIP box of p12 lacks the conserved glutamine; binds through a 2-fork plug made of an isoleucine and a tyrosine residue at +3 and +8 positions, respectively; and is stabilized by an aspartate at +6 position, which creates a network of intramolecular hydrogen bonds. These findings add to growing evidence that PCNA can bind a diverse range of protein sequences that may be broadly grouped as PIP-like motifs as has been previously suggested.
- From the CIC bioGUNE, Parque Tecnológico de Bizkaia Edificio 800, 48160 Derio, Spain.
Organizational Affiliation: