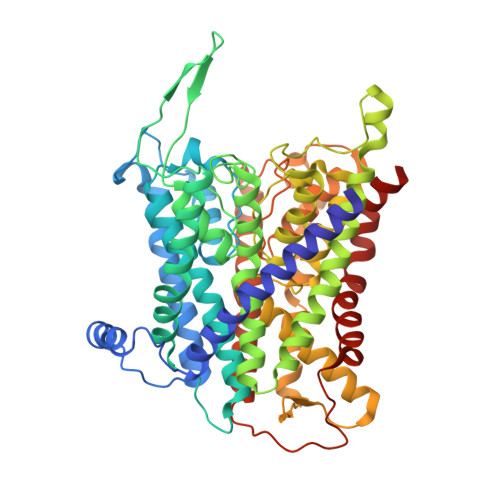
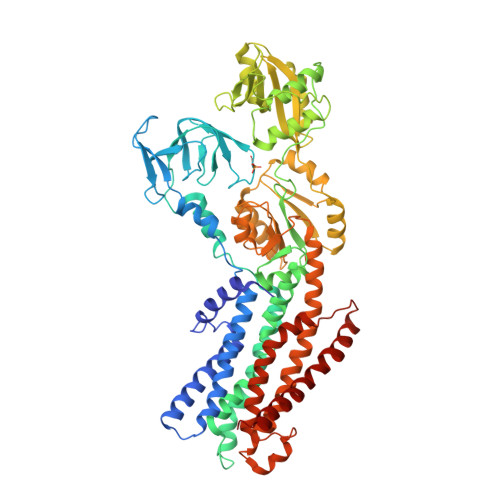
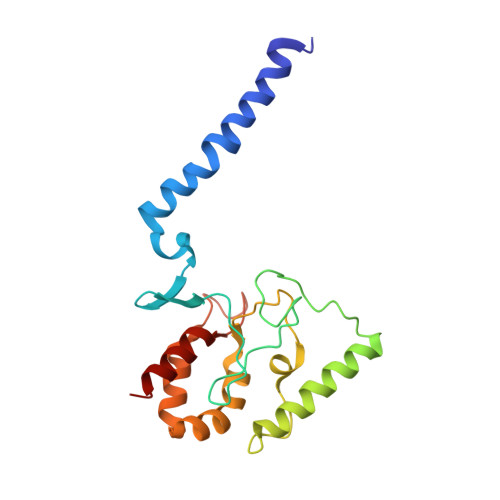
Cryo-EM structures of KdpFABC suggest a K+transport mechanism via two inter-subunit half-channels.
Stock, C., Hielkema, L., Tascon, I., Wunnicke, D., Oostergetel, G.T., Azkargorta, M., Paulino, C., Hanelt, I.(2018) Nat Commun 9: 4971-4971
- PubMed: 30478378
- DOI: https://doi.org/10.1038/s41467-018-07319-2
- Primary Citation of Related Structures:
6HRA, 6HRB - PubMed Abstract:
P-type ATPases ubiquitously pump cations across biological membranes to maintain vital ion gradients. Among those, the chimeric K + uptake system KdpFABC is unique. While ATP hydrolysis is accomplished by the P-type ATPase subunit KdpB, K + has been assumed to be transported by the channel-like subunit KdpA. A first crystal structure uncovered its overall topology, suggesting such a spatial separation of energizing and transporting units. Here, we report two cryo-EM structures of the 157 kDa, asymmetric KdpFABC complex at 3.7 Å and 4.0 Å resolution in an E1 and an E2 state, respectively. Unexpectedly, the structures suggest a translocation pathway through two half-channels along KdpA and KdpB, uniting the alternating-access mechanism of actively pumping P-type ATPases with the high affinity and selectivity of K + channels. This way, KdpFABC would function as a true chimeric complex, synergizing the best features of otherwise separately evolved transport mechanisms.
- Institute of Biochemistry, Biocenter, Goethe University Frankfurt, Max-von-Laue-Straße 9, 60438, Frankfurt/Main, Germany.
Organizational Affiliation: