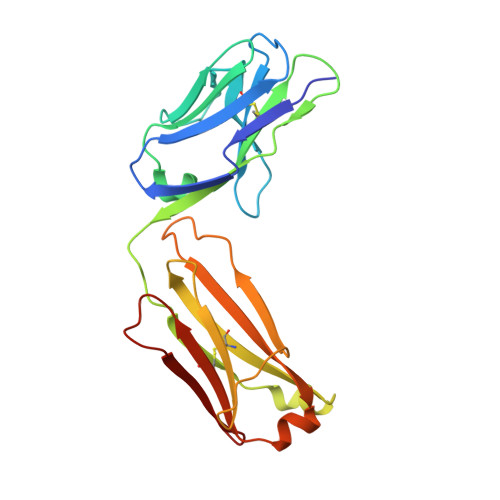
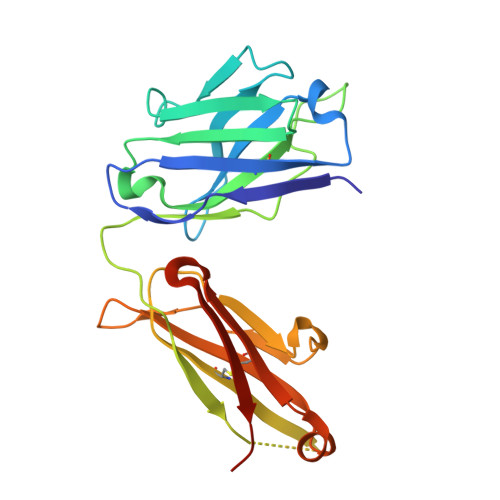
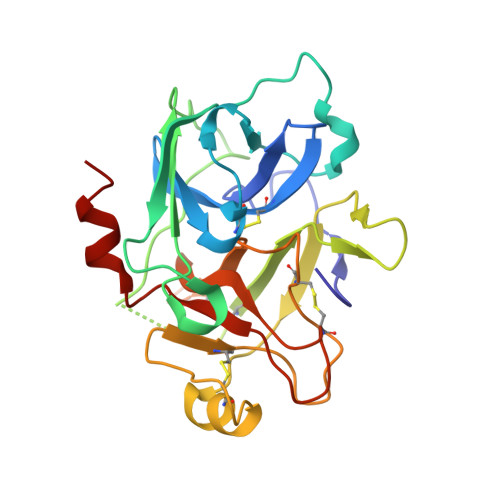
Allosteric Inhibition as a New Mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated Form of Coagulation Factor XI.
Schaefer, M., Buchmueller, A., Dittmer, F., Strassburger, J., Wilmen, A.(2019) J Mol Biology 431: 4817-4833
- PubMed: 31655039
- DOI: https://doi.org/10.1016/j.jmb.2019.09.008
- Primary Citation of Related Structures:
6HHC - PubMed Abstract:
Factor XI (FXI), the zymogen of activated FXI (FXIa), is an attractive target for novel anticoagulants because FXI inhibition offers the potential to reduce thrombosis risk while minimizing the risk of bleeding. BAY 1213790, a novel anti-FXIa antibody, was generated using phage display technology. Crystal structure analysis of the FXIa-BAY 1213790 complex demonstrated that the tyrosine-rich complementarity-determining region 3 loop of the heavy chain of BAY 1213790 penetrated deepest into the FXIa binding epitope, forming a network of favorable interactions including a direct hydrogen bond from Tyr102 to the Gln451 sidechain (2.9 Å). The newly discovered binding epitope caused a structural rearrangement of the FXIa active site, revealing a novel allosteric mechanism of FXIa inhibition by BAY 1213790. BAY 1213790 specifically inhibited FXIa with a binding affinity of 2.4 nM, and in human plasma, prolonged activated partial thromboplastin time and inhibited thrombin generation in a concentration-dependent manner.
- Bayer AG, Research and Development, Pharmaceuticals, Structural Biology, 13342 Berlin, Germany. Electronic address: martina.schaefer1@bayer.com.
Organizational Affiliation: