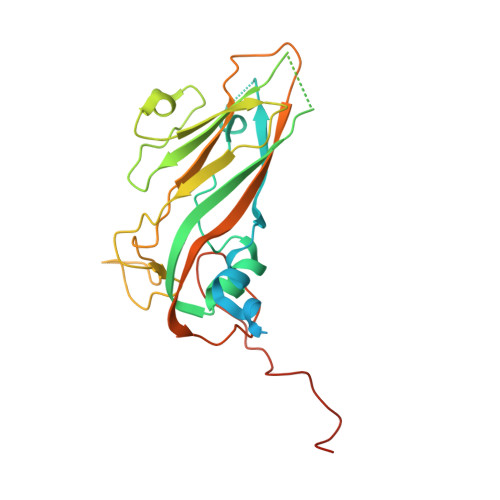
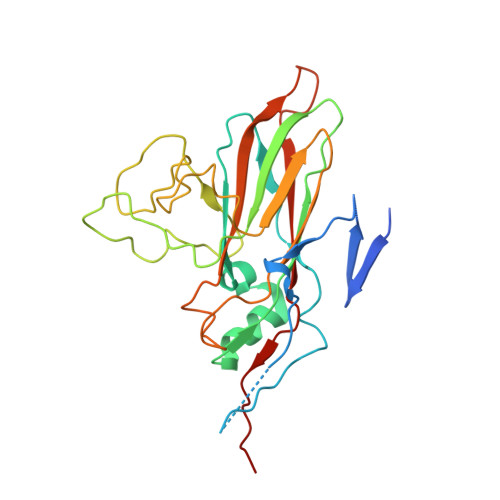
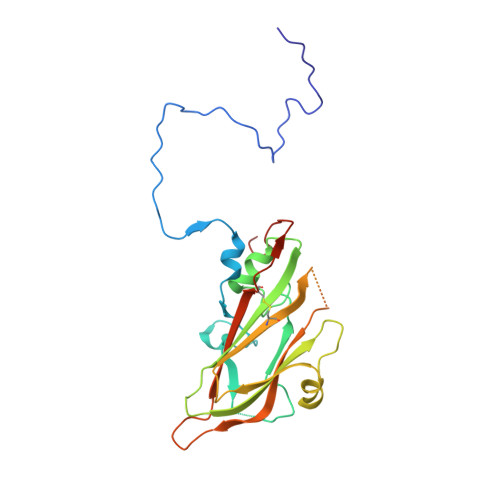
Enterovirus particles expel capsid pentamers to enable genome release.
Buchta, D., Fuzik, T., Hrebik, D., Levdansky, Y., Sukenik, L., Mukhamedova, L., Moravcova, J., Vacha, R., Plevka, P.(2019) Nat Commun 10: 1138-1138
- PubMed: 30850609
- DOI: https://doi.org/10.1038/s41467-019-09132-x
- Primary Citation of Related Structures:
6HBG, 6HBH, 6HBJ, 6HBK, 6HBL, 6HHT - PubMed Abstract:
Viruses from the genus Enterovirus are important human pathogens. Receptor binding or exposure to acidic pH in endosomes converts enterovirus particles to an activated state that is required for genome release. However, the mechanism of enterovirus uncoating is not well understood. Here, we use cryo-electron microscopy to visualize virions of human echovirus 18 in the process of genome release. We discover that the exit of the RNA from the particle of echovirus 18 results in a loss of one, two, or three adjacent capsid-protein pentamers. The opening in the capsid, which is more than 120 Å in diameter, enables the release of the genome without the need to unwind its putative double-stranded RNA segments. We also detect capsids lacking pentamers during genome release from echovirus 30. Thus, our findings uncover a mechanism of enterovirus genome release that could become target for antiviral drugs.
- Central European Institute of Technology, Masaryk University, Kamenice 5, Brno, 625 00, Czech Republic.
Organizational Affiliation: