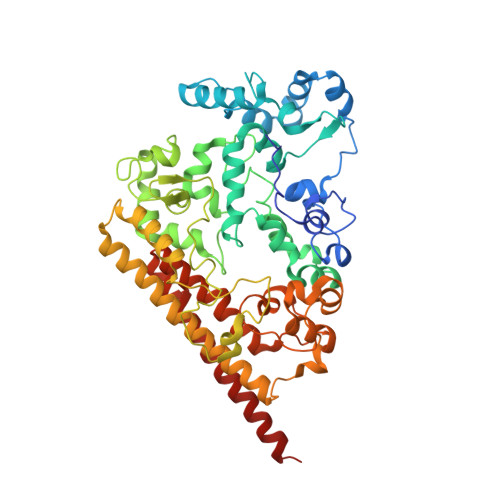
A 60-heme reductase complex from an anammox bacterium shows an extended electron transfer pathway.
Dietl, A., Maalcke, W.J., Ferousi, C., Jetten, M.S.M., Kartal, B., Barends, T.R.M.(2019) Acta Crystallogr D Struct Biol 75: 333-341
- PubMed: 30950404
- DOI: https://doi.org/10.1107/S2059798318017473
- Primary Citation of Related Structures:
6H5L - PubMed Abstract:
The hydroxylamine oxidoreductase/hydrazine dehydrogenase (HAO/HDH) protein family constitutes an important group of octaheme cytochromes c (OCCs). The majority of these proteins form homotrimers, with their subunits being covalently attached to each other via a rare cross-link between the catalytic heme moiety and a conserved tyrosine residue in an adjacent subunit. This covalent cross-link has been proposed to modulate the active-site heme towards oxidative catalysis by distorting the heme plane. In this study, the crystal structure of a stable complex of an HAO homologue (KsHAOr) with its diheme cytochrome c redox partner (KsDH) from the anammox bacterium Kuenenia stuttgartiensis was determined. KsHAOr lacks the tyrosine cross-link and is therefore tuned to reductive catalysis. The molecular model of the KsHAOr-KsDH complex at 2.6 Å resolution shows a heterododecameric (α 6 β 6 ) assembly, which was also shown to be the oligomeric state in solution by analytical ultracentrifugation and multi-angle static light scattering. The 60-heme-containing protein complex reveals a unique extended electron transfer pathway and provides deeper insights into catalysis and electron transfer in reductive OCCs.
- Department of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Jahnstrasse 29, 69120 Heidelberg, Germany.
Organizational Affiliation: