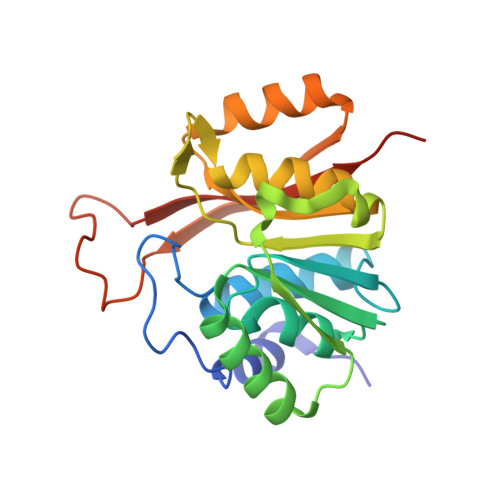
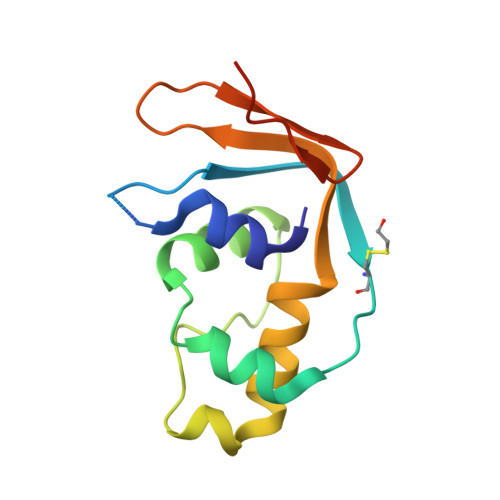
The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112.
van Tran, N., Ernst, F.G.M., Hawley, B.R., Zorbas, C., Ulryck, N., Hackert, P., Bohnsack, K.E., Bohnsack, M.T., Jaffrey, S.R., Graille, M., Lafontaine, D.L.J.(2019) Nucleic Acids Res 47: 7719-7733
- PubMed: 31328227
- DOI: https://doi.org/10.1093/nar/gkz619
- Primary Citation of Related Structures:
6H2U, 6H2V - PubMed Abstract:
N6-methyladenosine (m6A) has recently been found abundantly on messenger RNA and shown to regulate most steps of mRNA metabolism. Several important m6A methyltransferases have been described functionally and structurally, but the enzymes responsible for installing one m6A residue on each subunit of human ribosomes at functionally important sites have eluded identification for over 30 years. Here, we identify METTL5 as the enzyme responsible for 18S rRNA m6A modification and confirm ZCCHC4 as the 28S rRNA modification enzyme. We show that METTL5 must form a heterodimeric complex with TRMT112, a known methyltransferase activator, to gain metabolic stability in cells. We provide the first atomic resolution structure of METTL5-TRMT112, supporting that its RNA-binding mode differs distinctly from that of other m6A RNA methyltransferases. On the basis of similarities with a DNA methyltransferase, we propose that METTL5-TRMT112 acts by extruding the adenosine to be modified from a double-stranded nucleic acid.
- BIOC, CNRS, Ecole polytechnique, Institut Polytechnique de Paris, F-91128 Palaiseau, France.
Organizational Affiliation: