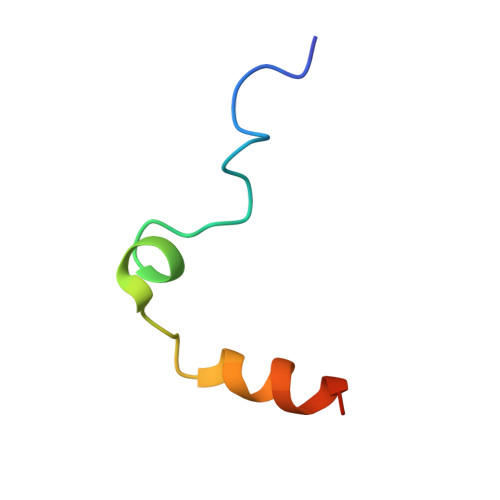
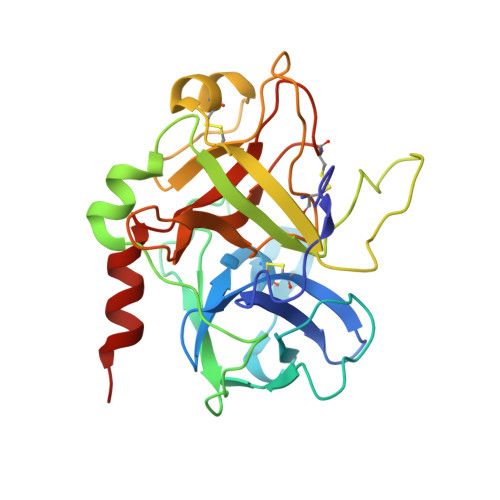
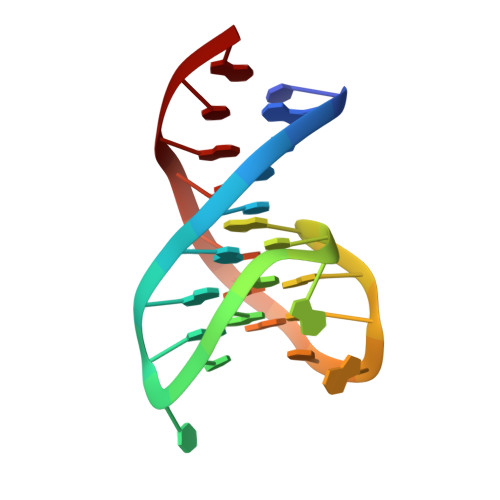
Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Troisi, R., Napolitano, V., Spiridonova, V., Russo Krauss, I., Sica, F.(2018) Nucleic Acids Res 46: 12177-12185
- PubMed: 30357392
- DOI: https://doi.org/10.1093/nar/gky990
- Primary Citation of Related Structures:
6EVV, 6GN7 - PubMed Abstract:
Despite aptamers are very promising alternative to antibodies, very few of them are under clinical trials or are used as drugs. Among them, NU172 is currently in Phase II as anticoagulant in heart disease treatments. It inhibits thrombin activity much more effectively than TBA, the best-known thrombin binding aptamer. The crystal structure of thrombin-NU172 complex reveals a bimodular duplex/quadruplex architecture for the aptamer, which binds thrombin exosite I through a highly complementary surface involving all three loops of the G-quadruplex module. Although the duplex domain does not interact directly with thrombin, the features of the duplex/quadruplex junction and the solution data on two newly designed NU172 mutants indicate that the duplex moiety is important for the optimization of the protein-ligand interaction and for the inhibition of the enzyme activity. Our work discloses the structural features determining the inhibition of thrombin by NU172 and put the basis for the design of mutants with improved properties.
- Department of Chemical Sciences, University of Naples 'Federico II', Naples 80126, Italy.
Organizational Affiliation: