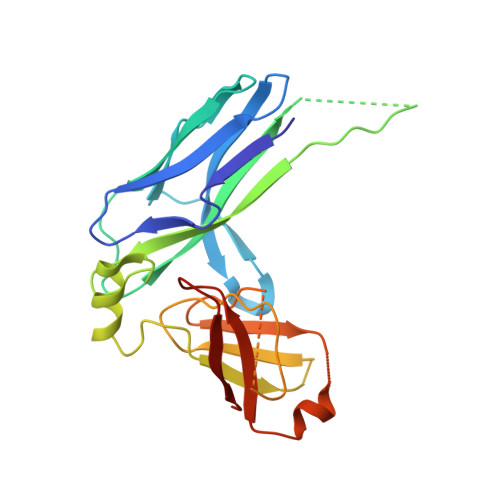
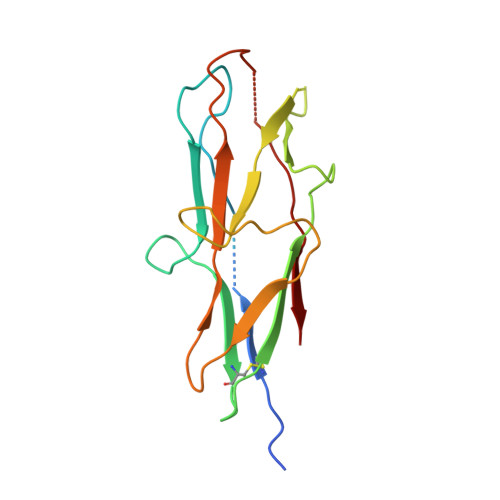
Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
Pakharukova, N., McKenna, S., Tuittila, M., Paavilainen, S., Malmi, H., Xu, Y., Parilova, O., Matthews, S., Zavialov, A.V.(2018) J Biological Chem 293: 17070-17080
- PubMed: 30228191
- DOI: https://doi.org/10.1074/jbc.RA118.004170
- Primary Citation of Related Structures:
6FM5, 6FQ0, 6FQA - PubMed Abstract:
Adhesive pili are external component of fibrous adhesive organelles and help bacteria attach to biotic or abiotic surfaces. The biogenesis of adhesive pili via the chaperone-usher pathway (CUP) is independent of external energy sources. In the classical CUP, chaperones transport assembly-competent pilins in a folded but expanded conformation. During donor-strand exchange, pilins subsequently collapse, producing a tightly packed hydrophobic core and releasing the necessary free energy to drive fiber formation. Here, we show that pilus biogenesis in non-classical, archaic, and alternative CUPs uses a different source of conformational energy. High-resolution structures of the archaic Csu-pili system from Acinetobacter baumannii revealed that non-classical chaperones employ a short donor strand motif that is insufficient to fully complement the pilin fold. This results in chaperone-bound pilins being trapped in a substantially unfolded intermediate. The exchange of this short motif with the longer donor strand from adjacent pilin provides the full steric information essential for folding, and thereby induces a large unfolded-to-folded conformational transition to drive assembly. Our findings may inform the development of anti-adhesion drugs (pilicides) to combat bacterial infections.
- From the Department of Chemistry, University of Turku, Joint Biotechnology Laboratory (JBL), Arcanum, Vatselankatu 2, Turku FIN-20500, Finland and.
Organizational Affiliation: