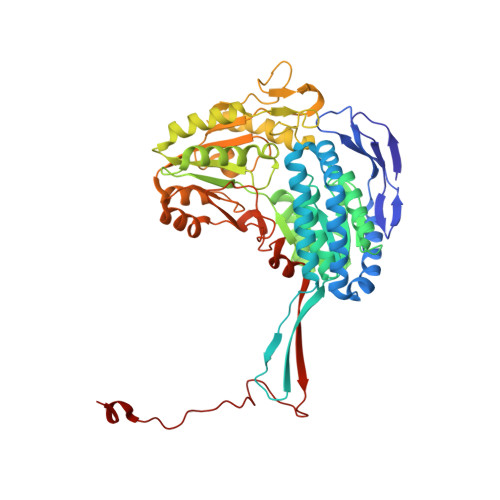
The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Hayes, K., Noor, M., Djeghader, A., Armshaw, P., Pembroke, T., Tofail, S., Soulimane, T.(2018) Sci Rep 8: 13327-13327
- PubMed: 30190503
- DOI: https://doi.org/10.1038/s41598-018-31724-8
- Primary Citation of Related Structures:
6FJX, 6FK3, 6FKU, 6FKV - PubMed Abstract:
Aldehyde dehydrogenases (ALDH) form a superfamily of dimeric or tetrameric enzymes that catalyze the oxidation of a broad range of aldehydes into their corresponding carboxylic acids with the concomitant reduction of the cofactor NAD(P) into NAD(P)H. Despite their varied polypeptide chain length and oligomerisation states, ALDHs possess a conserved architecture of three domains: the catalytic domain, NAD(P) + binding domain, and the oligomerization domain. Here, we describe the structure and function of the ALDH from Thermus thermophilus (ALDH Tt ) which exhibits non-canonical features of both dimeric and tetrameric ALDH and a previously uncharacterized C-terminal arm extension forming novel interactions with the N-terminus in the quaternary structure. This unusual tail also interacts closely with the substrate entry tunnel in each monomer providing further mechanistic detail for the recent discovery of tail-mediated activity regulation in ALDH. However, due to the novel distal extension of the tail of ALDH Tt and stabilizing termini-interactions, the current model of tail-mediated substrate access is not apparent in ALDH Tt . The discovery of such a long tail in a deeply and early branching phylum such as Deinococcus-Thermus indicates that ALDH Tt may be an ancestral or primordial metabolic model of study. This structure provides invaluable evidence of how metabolic regulation has evolved and provides a link to early enzyme regulatory adaptations.
- Department of Chemical Sciences, University of Limerick, Limerick, V94 T9PX, Ireland.
Organizational Affiliation: