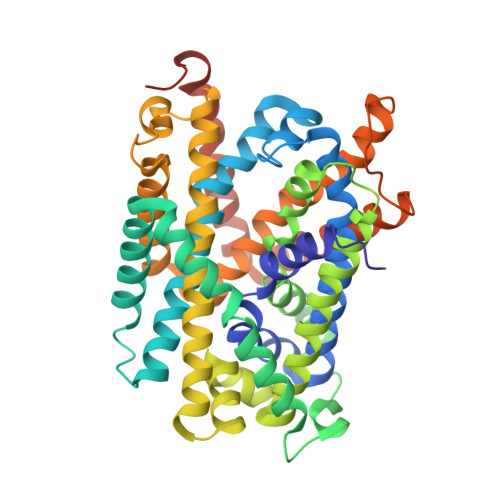
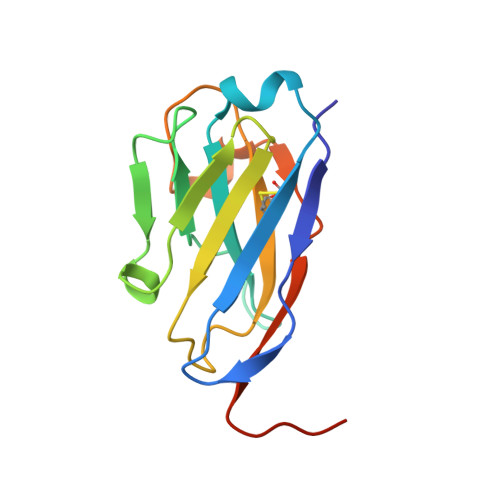
L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Errasti-Murugarren, E., Fort, J., Bartoccioni, P., Diaz, L., Pardon, E., Carpena, X., Espino-Guarch, M., Zorzano, A., Ziegler, C., Steyaert, J., Fernandez-Recio, J., Fita, I., Palacin, M.(2019) Nat Commun 10: 1807-1807
- PubMed: 31000719
- DOI: https://doi.org/10.1038/s41467-019-09837-z
- Primary Citation of Related Structures:
6F2G, 6F2W - PubMed Abstract:
L-amino acid transporters (LATs) play key roles in human physiology and are implicated in several human pathologies. LATs are asymmetric amino acid exchangers where the low apparent affinity cytoplasmic side controls the exchange of substrates with high apparent affinity on the extracellular side. Here, we report the crystal structures of an LAT, the bacterial alanine-serine-cysteine exchanger (BasC), in a non-occluded inward-facing conformation in both apo and substrate-bound states. We crystallized BasC in complex with a nanobody, which blocks the transporter from the intracellular side, thus unveiling the sidedness of the substrate interaction of BasC. Two conserved residues in human LATs, Tyr 236 and Lys 154, are located in equivalent positions to the Na1 and Na2 sites of sodium-dependent APC superfamily transporters. Functional studies and molecular dynamics (MD) calculations reveal that these residues are key for the asymmetric substrate interaction of BasC and in the homologous human transporter Asc-1.
- Institute for Research in Biomedicine (IRB Barcelona), Barcelona Institute of Science and Technology, 08028, Barcelona, Spain.
Organizational Affiliation: