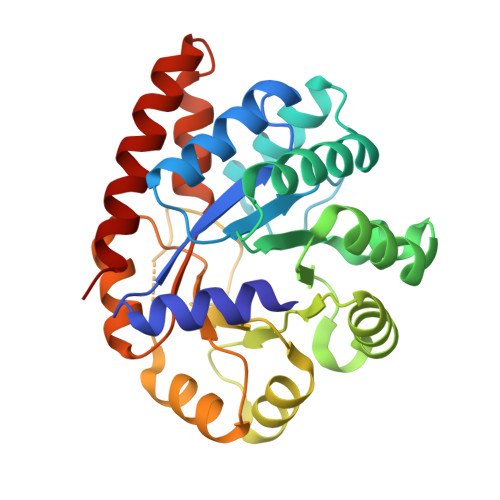
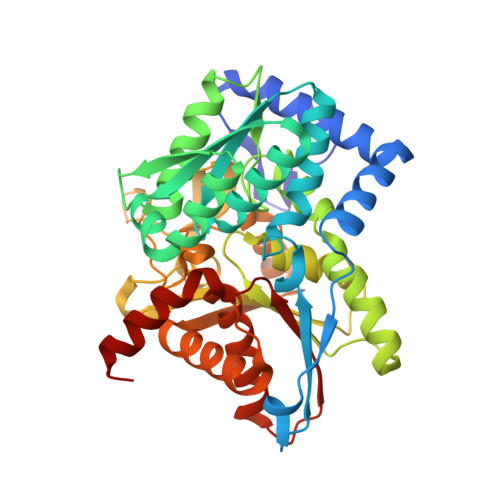
Tryptophan synthase Q114A mutant in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate (P1T) at the beta site, and cesium ion at the metal coordination site.
Hilario, E., Dunn, M.F., Mueller, L.J., Fan, L.To be published.