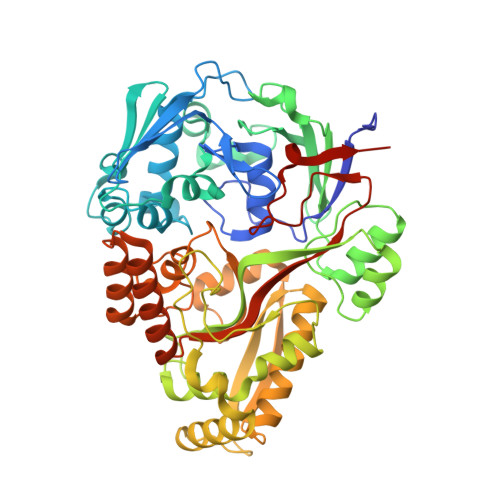
Oligopeptide-binding protein from nontypeableHaemophilus influenzaehas ligand-specific sites to accommodate peptides and heme in the binding pocket.
Tanaka, K.J., Pinkett, H.W.(2019) J Biological Chem 294: 1070-1082
- PubMed: 30455346
- DOI: https://doi.org/10.1074/jbc.RA118.004479
- Primary Citation of Related Structures:
6DQQ, 6DQR, 6DQT, 6DQU, 6DTF, 6DTG, 6DTH - PubMed Abstract:
In nontypeable Haemophilus influenzae (NTHi), the oligopeptide-binding protein (OppA) serves as the substrate-binding protein (SBP) of the oligopeptide transport system responsible for the import of peptides. We solved the crystal structure of nthiOppA in complex with hydrophobic peptides of various sizes. Our novel hexapeptide complex demonstrates the flexibility of the nthiOppA-binding cavity to expand and accommodate the longer peptide while maintaining similar protein-peptide interactions of smaller peptide complexes. In addition to acquiring peptides from the host environment, as a heme auxotroph NTHi utilizes host hemoproteins as a source of essential iron. OppA is a member of the Cluster C SBP family, and unlike other SBP families, some members recognize two distinctly different substrates. DppA (dipeptide), MppA (murein tripeptide), and SapA (antimicrobial peptides) are Cluster C proteins known to also transport heme. We observed nthiOppA shares this heme-binding characteristic and established heme specificity and affinity by surface plasmon resonance (SPR) of the four Cluster C proteins in NTHi. Ligand-docking studies predicted a distinct heme-specific cleft in the binding pocket, and using SPR competition assays, we observed that heme does not directly compete with peptide in the substrate-binding pocket. Additionally, we identified that the individual nthiOppA domains differentially contribute to substrate binding, with one domain playing a dominant role in heme binding and the other in peptide binding. Our results demonstrate the multisubstrate specificity of nthiOppA and the role of NTHi Cluster C proteins in the heme-uptake pathway for this pathogen.
- From the Department of Molecular Biosciences, Northwestern University, Evanston, Illinois 60208.
Organizational Affiliation: