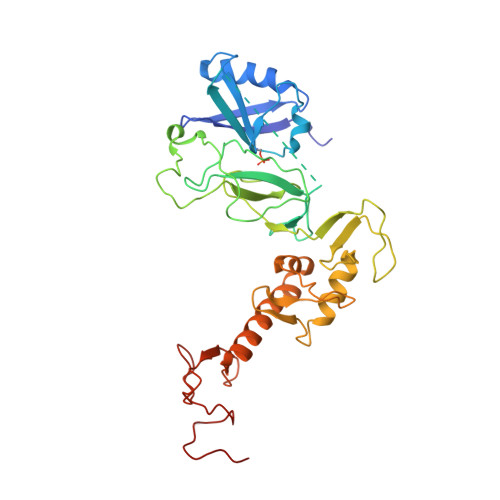
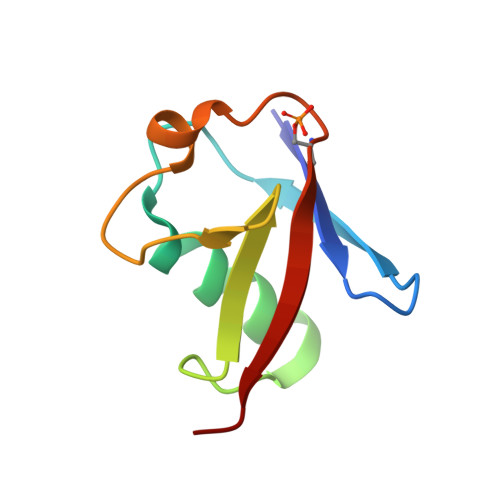
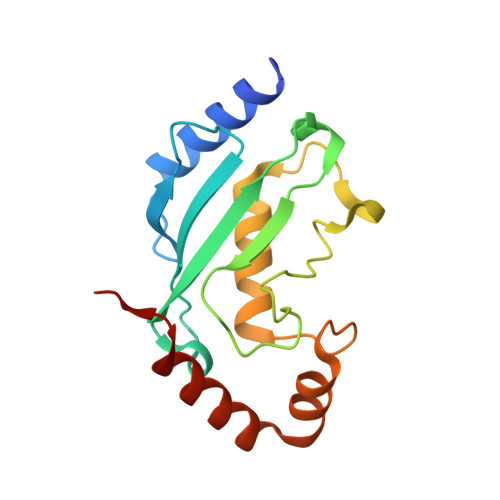
Mechanism of parkin activation by phosphorylation.
Sauve, V., Sung, G., Soya, N., Kozlov, G., Blaimschein, N., Miotto, L.S., Trempe, J.F., Lukacs, G.L., Gehring, K.(2018) Nat Struct Mol Biol 25: 623-630
- PubMed: 29967542
- DOI: https://doi.org/10.1038/s41594-018-0088-7
- Primary Citation of Related Structures:
6DJW, 6DJX - PubMed Abstract:
Mutations in the ubiquitin ligase parkin are responsible for a familial form of Parkinson's disease. Parkin and the PINK1 kinase regulate a quality-control system for mitochondria. PINK1 phosphorylates ubiquitin on the outer membrane of damaged mitochondria, thus leading to recruitment and activation of parkin via phosphorylation of its ubiquitin-like (Ubl) domain. Here, we describe the mechanism of parkin activation by phosphorylation. The crystal structure of phosphorylated Bactrocera dorsalis (oriental fruit fly) parkin in complex with phosphorylated ubiquitin and an E2 ubiquitin-conjugating enzyme reveals that the key activating step is movement of the Ubl domain and release of the catalytic RING2 domain. Hydrogen/deuterium exchange and NMR experiments with the various intermediates in the activation pathway confirm and extend the interpretation of the crystal structure to mammalian parkin. Our results rationalize previously unexplained Parkinson's disease mutations and the presence of internal linkers that allow large domain movements in parkin.
- Department of Biochemistry, McGill University, Montreal, Quebec, Canada.
Organizational Affiliation: