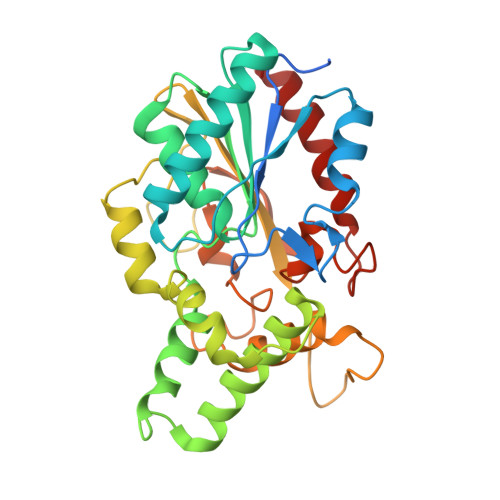
Structure solution and analyses of the first true lipase obtained from metagenomics indicate potential for increased thermostability.
Martini, V.P., Krieger, N., Glogauer, A., Souza, E.M., Iulek, J.(2019) N Biotechnol 53: 65-72
- PubMed: 31306784
- DOI: https://doi.org/10.1016/j.nbt.2019.07.001
- Primary Citation of Related Structures:
6CL4 - PubMed Abstract:
Metagenomics is a modern approach to discovery of new enzymes with novel properties. This article reports the structure of a new lipase, belonging to family I.1, obtained by means of metagenomics. Its structure presents a fold typical of α/β hydrolases, with the lid in closed conformation. The protein was previously shown to present high thermostability and to be stable in aqueous solutions of polar organic solvents at high concentrations [30% (V/V)]. Molecular dynamics studies showed that the protein maintains its structure well in organic solvents. They also suggested that its thermostability might be enhanced if it were mutated to present a disulfide bond similar to that typically found in lipase family I.2. These findings identify this lipase as a good candidate for further improvement through protein engineering.
- Department of Chemistry, Federal University of Paraná, Rua Francisco Heráclito dos Santos, 100, Curitiba, PR, 81531-990, Brazil. Electronic address: viviane.martini@ifpr.edu.br.
Organizational Affiliation: