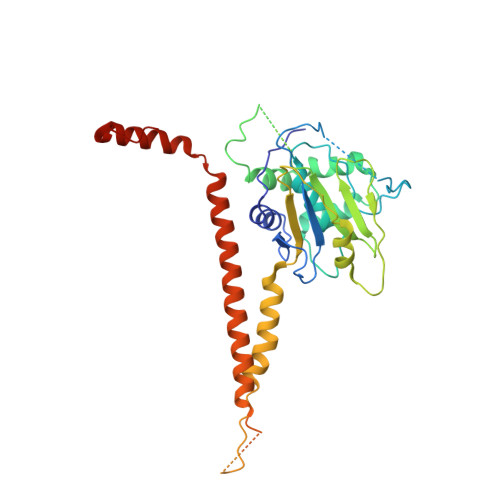
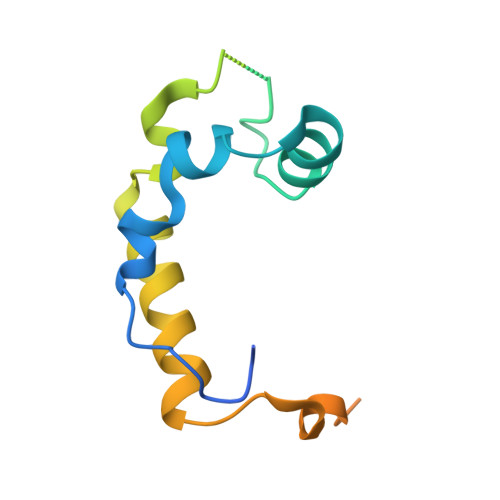
A bidentate Polycomb Repressive-Deubiquitinase complex is required for efficient activity on nucleosomes.
Foglizzo, M., Middleton, A.J., Burgess, A.E., Crowther, J.M., Dobson, R.C.J., Murphy, J.M., Day, C.L., Mace, P.D.(2018) Nat Commun 9: 3932-3932
- PubMed: 30258054
- DOI: https://doi.org/10.1038/s41467-018-06186-1
- Primary Citation of Related Structures:
6CGA - PubMed Abstract:
Attachment of ubiquitin to lysine 119 of Histone 2A (H2AK119Ub) is an epigenetic mark characteristic of repressed developmental genes, which is removed by the Polycomb Repressive-Deubiquitinase (PR-DUB) complex. Here we report the crystal structure of the Drosophila PR-DUB, revealing that the deubiquitinase Calypso and its activating partner ASX form a 2:2 complex. The bidentate Calypso-ASX complex is generated by dimerisation of two activated Calypso proteins through their coiled-coil regions. Disrupting the Calypso dimer interface does not affect inherent catalytic activity, but inhibits removal of H2AK119Ub as a consequence of impaired recruitment to nucleosomes. Mutating the equivalent surface on the human counterpart, BAP1, also compromises activity on nucleosomes. Together, this suggests that high local concentrations drive assembly of bidentate PR-DUB complexes on chromatin-providing a mechanistic basis for enhanced PR-DUB activity at specific genomic foci, and the impact of distinct classes of PR-DUB mutations in tumorigenesis.
- Biochemistry Department, School of Biomedical Sciences, University of Otago, P.O. Box 56, 710, Cumberland St., Dunedin, 9054, New Zealand.
Organizational Affiliation: