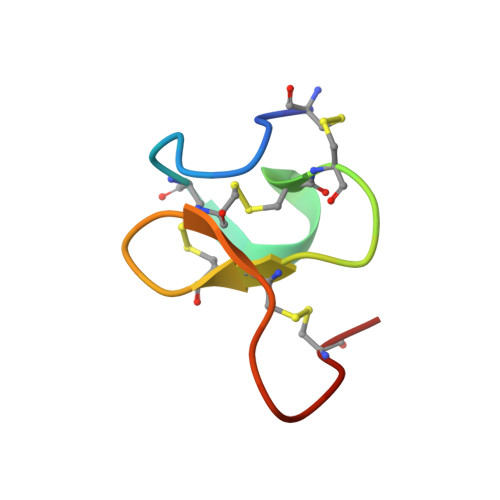
Chemical Synthesis and NMR Solution Structure of Conotoxin GXIA from Conus geographus .
Armstrong, D.A., Jin, A.H., Braga Emidio, N., Lewis, R.J., Alewood, P.F., Rosengren, K.J.(2021) Mar Drugs 19
- PubMed: 33530397
- DOI: https://doi.org/10.3390/md19020060
- Primary Citation of Related Structures:
6CEI - PubMed Abstract:
Conotoxins are disulfide-rich peptides found in the venom of cone snails. Due to their exquisite potency and high selectivity for a wide range of voltage and ligand gated ion channels they are attractive drug leads in neuropharmacology. Recently, cone snails were found to have the capability to rapidly switch between venom types with different proteome profiles in response to predatory or defensive stimuli. A novel conotoxin, GXIA (original name G117), belonging to the I 3 -subfamily was identified as the major component of the predatory venom of piscivorous Conus geographus . Using 2D solution NMR spectroscopy techniques, we resolved the 3D structure for GXIA, the first structure reported for the I 3 -subfamily and framework XI family. The 32 amino acid peptide is comprised of eight cysteine residues with the resultant disulfide connectivity forming an ICK+1 motif. With a triple stranded β-sheet, the GXIA backbone shows striking similarity to several tarantula toxins targeting the voltage sensor of voltage gated potassium and sodium channels. Supported by an amphipathic surface, the structural evidence suggests that GXIA is able to embed in the membrane and bind to the voltage sensor domain of a putative ion channel target.
- School of Biomedical Sciences, Faculty of Medicine, The University of Queensland, Brisbane, QLD 4072, Australia.
Organizational Affiliation: