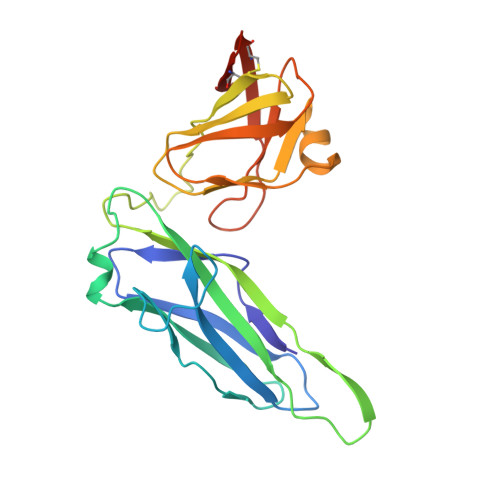
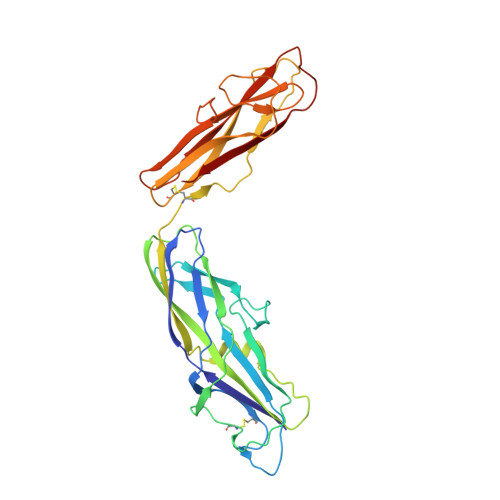
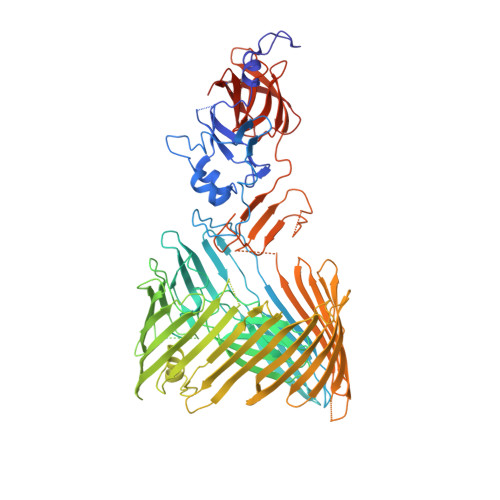
Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Omattage, N.S., Deng, Z., Pinkner, J.S., Dodson, K.W., Almqvist, F., Yuan, P., Hultgren, S.J.(2018) Nat Microbiol 3: 1362-1368
- PubMed: 30275511
- DOI: https://doi.org/10.1038/s41564-018-0255-y
- Primary Citation of Related Structures:
6CD2 - PubMed Abstract:
Chaperone-usher pathway pili are extracellular proteinaceous fibres ubiquitously found on Gram-negative bacteria, and mediate host-pathogen interactions and biofilm formation critical in pathogenesis in numerous human diseases 1 . During pilus assembly, an outer membrane macromolecular machine called the usher catalyses pilus biogenesis from the individual subunits that are delivered as chaperone-subunit complexes in the periplasm. The usher orchestrates pilus assembly using all five functional domains: a 24-stranded transmembrane β-barrel translocation domain, a β-sandwich plug domain, an amino-terminal periplasmic domain and two carboxy-terminal periplasmic domains (CTD1 and CTD2) 2-6 . Despite extensive structural and functional characterization, the mechanism by which the usher is activated to initiate pilus biogenesis is unknown. Here, we present the crystal structure of the full-length PapC usher from Escherichia coli in complex with its cognate PapDG chaperone-subunit complex in a pre-activation state, elucidating molecular details of how the usher is specifically engaged by allosteric interactions with its substrate preceding activation and how the usher facilitates the transfer of subunits from the amino-terminal periplasmic domain to the CTDs during pilus assembly. This work elucidates the intricate workings of a molecular machine that catalyses chaperone-usher pathway pilus assembly and opens the door for the development of potent inhibitors to block pilus biogenesis.
- Department of Molecular Microbiology, Washington University in St Louis, St Louis, MO, USA.
Organizational Affiliation: