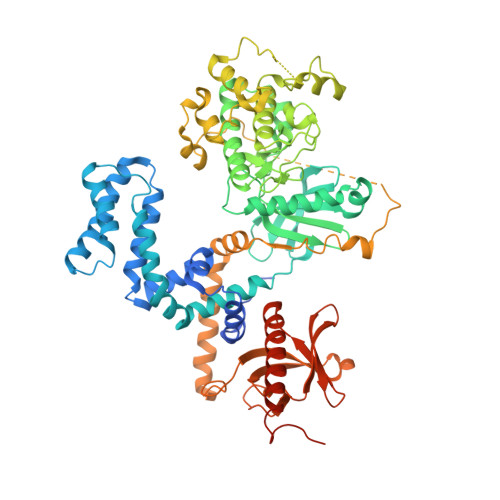
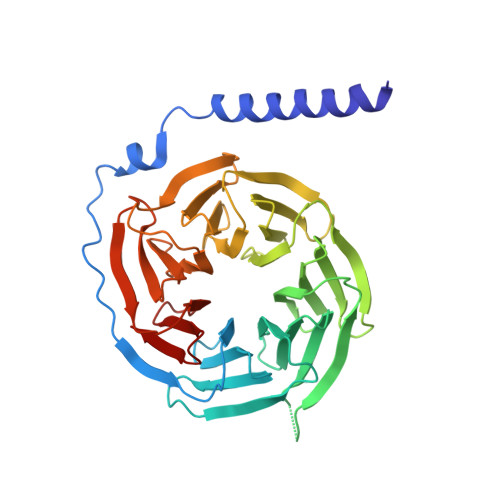
Utilizing a structure-based docking approach to develop potent G protein-coupled receptor kinase (GRK) 2 and 5 inhibitors.
Waldschmidt, H.V., Bouley, R., Kirchhoff, P.D., Lee, P., Tesmer, J.J.G., Larsen, S.D.(2018) Bioorg Med Chem Lett 28: 1507-1515
- PubMed: 29627263
- DOI: https://doi.org/10.1016/j.bmcl.2018.03.082
- Primary Citation of Related Structures:
6C2Y - PubMed Abstract:
G protein-coupled receptor (GPCR) kinases (GRKs) regulate the desensitization and internalization of GPCRs. Two of these, GRK2 and GRK5, are upregulated in heart failure and are promising targets for heart failure treatment. Although there have been several reports of potent and selective inhibitors of GRK2 there are few for GRK5. Herein, we describe a ligand docking approach utilizing the crystal structures of the GRK2-Gβγ·GSK180736A and GRK5·CCG215022 complexes to search for amide substituents predicted to confer GRK2 and/or GRK5 potency and selectivity. From this campaign, we successfully generated two new potent GRK5 inhibitors, although neither exhibited selectivity over GRK2.
- Department of Medicinal Chemistry, College of Pharmacy, University of Michigan, Ann Arbor, MI, United States; Vahlteich Medicinal Chemistry Core, College of Pharmacy, University of Michigan, Ann Arbor, MI, United States.
Organizational Affiliation: