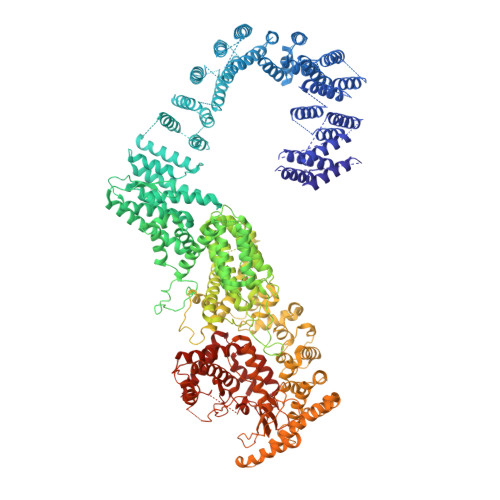
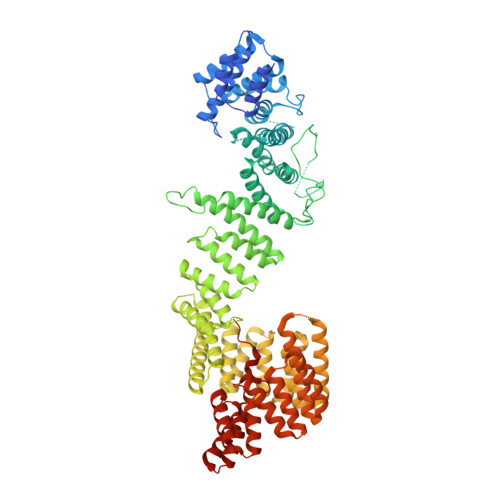
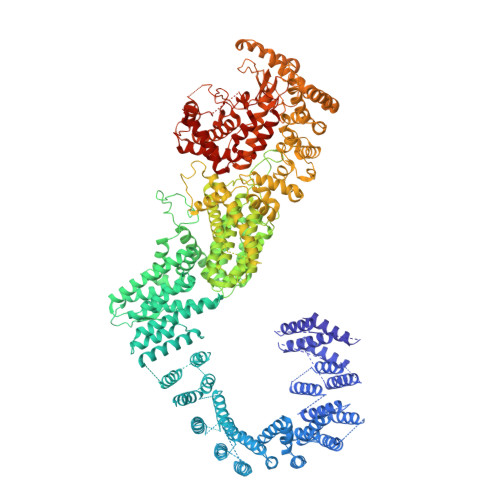
Architecture of the human PI4KIII alpha lipid kinase complex.
Lees, J.A., Zhang, Y., Oh, M.S., Schauder, C.M., Yu, X., Baskin, J.M., Dobbs, K., Notarangelo, L.D., De Camilli, P., Walz, T., Reinisch, K.M.(2017) Proc Natl Acad Sci U S A 114: 13720-13725
- PubMed: 29229838
- DOI: https://doi.org/10.1073/pnas.1718471115
- Primary Citation of Related Structures:
6BQ1 - PubMed Abstract:
Plasma membrane (PM) phosphoinositides play essential roles in cell physiology, serving as both markers of membrane identity and signaling molecules central to the cell's interaction with its environment. The first step in PM phosphoinositide synthesis is the conversion of phosphatidylinositol (PI) to PI4P, the precursor of PI(4,5)P 2 and PI(3,4,5)P 3 This conversion is catalyzed by the PI4KIIIα complex, comprising a lipid kinase, PI4KIIIα, and two regulatory subunits, TTC7 and FAM126. We here report the structure of this complex at 3.6-Å resolution, determined by cryo-electron microscopy. The proteins form an obligate ∼700-kDa superassembly with a broad surface suitable for membrane interaction, toward which the kinase active sites are oriented. The structural complexity of the assembly highlights PI4P synthesis as a major regulatory junction in PM phosphoinositide homeostasis. Our studies provide a framework for further exploring the mechanisms underlying PM phosphoinositide regulation.
- Department of Cell Biology, Yale University School of Medicine, New Haven, CT 06520.
Organizational Affiliation: