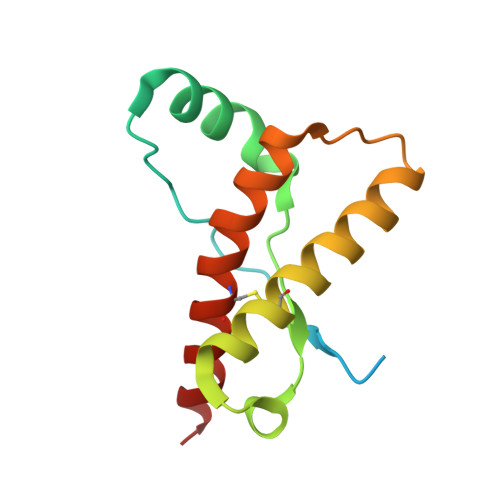
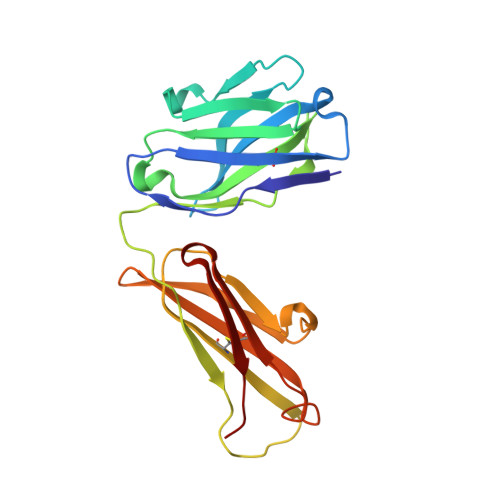
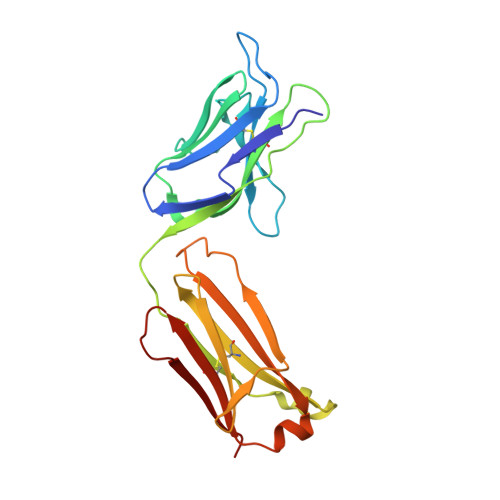
Structural characterization of POM6 Fab and mouse prion protein complex identifies key regions for prions conformational conversion.
Baral, P.K., Swayampakula, M., Aguzzi, A., James, M.N.G.(2018) FEBS J 285: 1701-1714
- PubMed: 29569342
- DOI: https://doi.org/10.1111/febs.14438
- Primary Citation of Related Structures:
6AQ7 - PubMed Abstract:
Conversion of the cellular prion protein PrP C into its pathogenic isoform PrP S c is the hallmark of prion diseases, fatal neurodegenerative diseases affecting many mammalian species including humans. Anti-prion monoclonal antibodies can arrest the progression of prion diseases by stabilizing the cellular form of the prion protein. Here, we present the crystal structure of the POM6 Fab fragment, in complex with the mouse prion protein (moPrP). The prion epitope of POM6 is in close proximity to the epitope recognized by the purportedly toxic antibody fragment, POM1 Fab also complexed with moPrP. The POM6 Fab recognizes a larger binding interface indicating a likely stronger binding compared to POM1. POM6 and POM1 exhibit distinct biological responses. Structural comparisons of the bound mouse prion proteins from the POM6 Fab:moPrP and POM1 Fab:moPrP complexes reveal several key regions of the prion protein that might be involved in initiating mis-folding events. The structural data of moPrP:POM6 Fab complex are available in the PDB under the accession number www.rcsb.org/pdb/search/structidSearch.do?structureId=6AQ7.
- Department of Biochemistry, Faculty of Medicine and Dentistry, University of Alberta, Edmonton, Canada.
Organizational Affiliation: