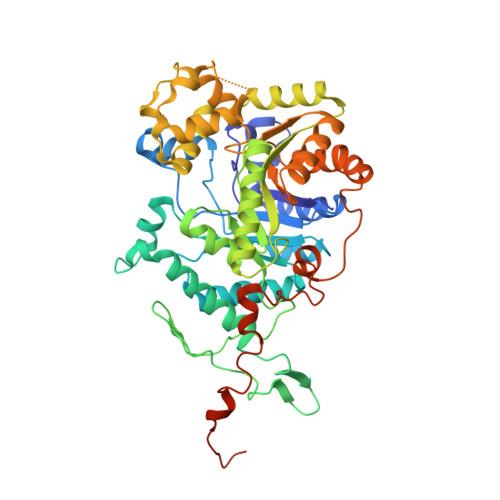
Crystal structure of the dimethylsulfide monooxygenase DmoA from Hyphomicrobium sulfonivorans.
Cao, H.Y., Wang, P., Peng, M., Shao, X., Chen, X.L., Li, C.Y.(2018) Acta Crystallogr F Struct Biol Commun 74: 781-786
- PubMed: 30511672
- DOI: https://doi.org/10.1107/S2053230X18015844
- Primary Citation of Related Structures:
6AK1 - PubMed Abstract:
DmoA is a monooxygenase which uses dioxygen (O 2 ) and reduced flavin mononucleotide (FMNH 2 ) to catalyze the oxidation of dimethylsulfide (DMS). Although it has been characterized, the structure of DmoA remains unknown. Here, the crystal structure of DmoA was determined to a resolution of 2.28 Å and was compared with those of its homologues LadA and BdsA. The results showed that their overall structures are similar: they all share a conserved TIM-barrel fold which is composed of eight α-helices and eight β-strands. In addition, they all have five additional insertions. Detailed comparison showed that the structures have notable differences despite their high sequence similarity. The substrate-binding pocket of DmoA is smaller compared with those of LadA and BdsA.
- Marine Biotechnology Research Center, State Key Laboratory of Microbial Technology, Shandong University, Qingdao 266237, People's Republic of China.
Organizational Affiliation: