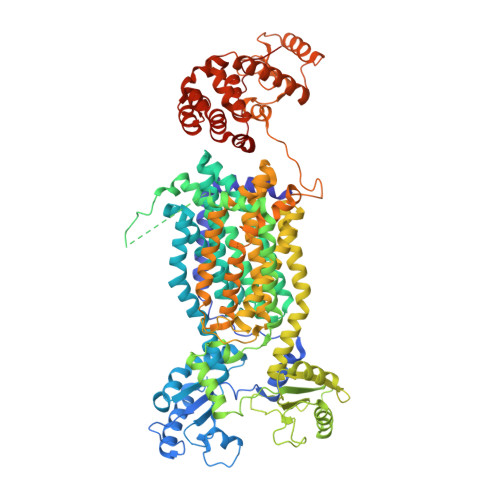
Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Zhang, B., Li, J., Yang, X., Wu, L., Zhang, J., Yang, Y., Zhao, Y., Zhang, L., Yang, X., Yang, X., Cheng, X., Liu, Z., Jiang, B., Jiang, H., Guddat, L.W., Yang, H., Rao, Z.(2019) Cell 176: 636-648.e13
- PubMed: 30682372
- DOI: https://doi.org/10.1016/j.cell.2019.01.003
- Primary Citation of Related Structures:
6AJF, 6AJG, 6AJH, 6AJI, 6AJJ - PubMed Abstract:
Despite intensive efforts to discover highly effective treatments to eradicate tuberculosis (TB), it remains as a major threat to global human health. For this reason, new TB drugs directed toward new targets are highly coveted. MmpLs (Mycobacterial membrane proteins Large), which play crucial roles in transporting lipids, polymers and immunomodulators and which also extrude therapeutic drugs, are among the most important therapeutic drug targets to emerge in recent times. Here, crystal structures of mycobacterial MmpL3 alone and in complex with four TB drug candidates, including SQ109 (in Phase 2b-3 clinical trials), are reported. MmpL3 consists of a periplasmic pore domain and a twelve-helix transmembrane domain. Two Asp-Tyr pairs centrally located in this domain appear to be key facilitators of proton-translocation. SQ109, AU1235, ICA38, and rimonabant bind inside the transmembrane region and disrupt these Asp-Tyr pairs. This structural data will greatly advance the development of MmpL3 inhibitors as new TB drugs.
- Shanghai Institute for Advanced Immunochemical Studies and School of Life Science and Technology, ShanghaiTech University, Shanghai, 201210, China; CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, 200031, China; University of Chinese Academy of Sciences, Beijing, 100101, China.
Organizational Affiliation: