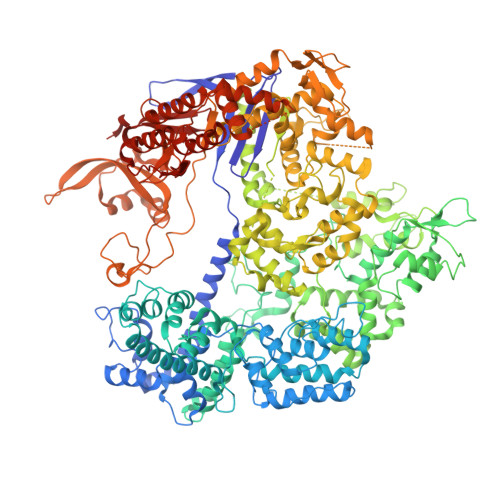
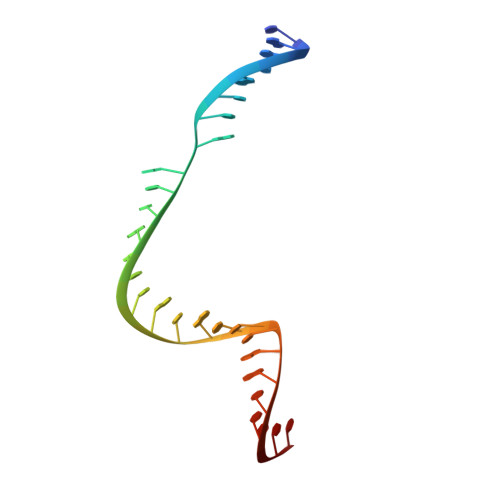
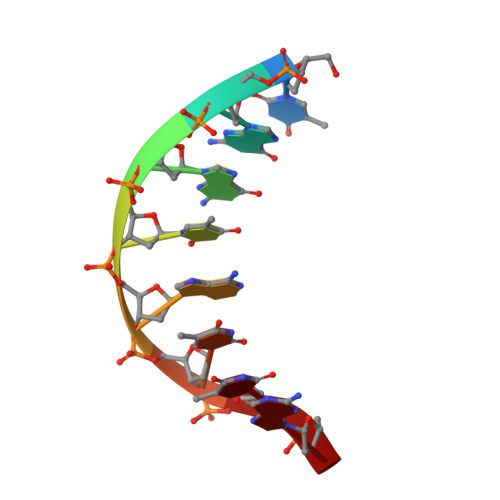
Engineered CRISPR-Cas9 nuclease with expanded targeting space
Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., Noda, T., Abudayyeh, O.O., Gootenberg, J.S., Mori, H., Oura, S., Holmes, B., Tanaka, M., Seki, M., Hirano, H., Aburatani, H., Ishitani, R., Ikawa, M., Yachie, N., Zhang, F., Nureki, O.(2018) Science 361: 1259-1262
- PubMed: 30166441
- DOI: https://doi.org/10.1126/science.aas9129
- Primary Citation of Related Structures:
6AI6 - PubMed Abstract:
The RNA-guided endonuclease Cas9 cleaves its target DNA and is a powerful genome-editing tool. However, the widely used Streptococcus pyogenes Cas9 enzyme (SpCas9) requires an NGG protospacer adjacent motif (PAM) for target recognition, thereby restricting the targetable genomic loci. Here, we report a rationally engineered SpCas9 variant (SpCas9-NG) that can recognize relaxed NG PAMs. The crystal structure revealed that the loss of the base-specific interaction with the third nucleobase is compensated by newly introduced non-base-specific interactions, thereby enabling the NG PAM recognition. We showed that SpCas9-NG induces indels at endogenous target sites bearing NG PAMs in human cells. Furthermore, we found that the fusion of SpCas9-NG and the activation-induced cytidine deaminase (AID) mediates the C-to-T conversion at target sites with NG PAMs in human cells.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. nisimasu@bs.s.u-tokyo.ac.jp nureki@bs.s.u-tokyo.ac.jp.
Organizational Affiliation: