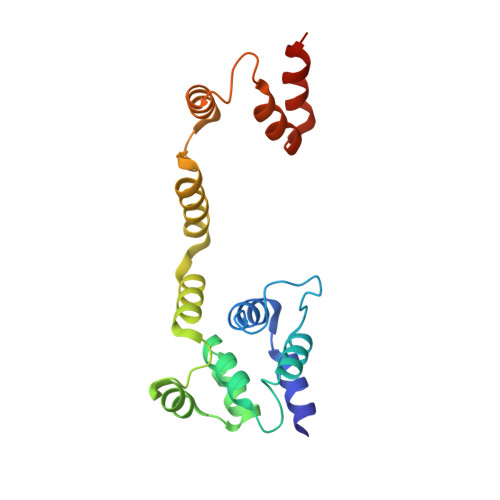
Molecular basis of dimerization of initiator caspase was revealed by crystal structure of caspase-8 pro-domain.
Park, H.H.(2019) Cell Death Differ 26: 1213-1220
- PubMed: 30206319
- DOI: https://doi.org/10.1038/s41418-018-0200-x
- Primary Citation of Related Structures:
6AGW - PubMed Abstract:
The assembly of death-inducing signaling complex (DISC) for activation of initiator caspase is a key step for the receptor-mediated apoptosis signaling. Many death effector domain (DED)-containing proteins are involved in DISC assembly and controlling. One of the main DISC component, caspase-8, contains DED and DED-mediated dimerization and oligomerization in the DISC is critical for the activation of this initiator caspase. There have been intensive studies to understand DED-mediated dimerization and oligomerization for the DISC assembly but no clear answer has been provided and there are many controversial arguments. Here, we suggested novel dimerization process of tandem DED of caspase-8 with crystallographic study.
- College of Pharmacy, Chung-Ang University, Seoul, 06974, South Korea. xrayleox@cau.ac.kr.
Organizational Affiliation: