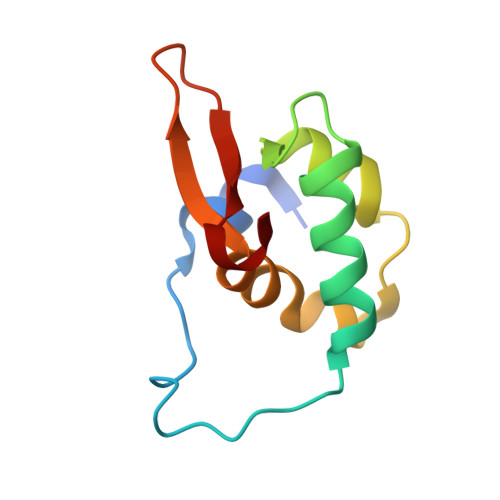
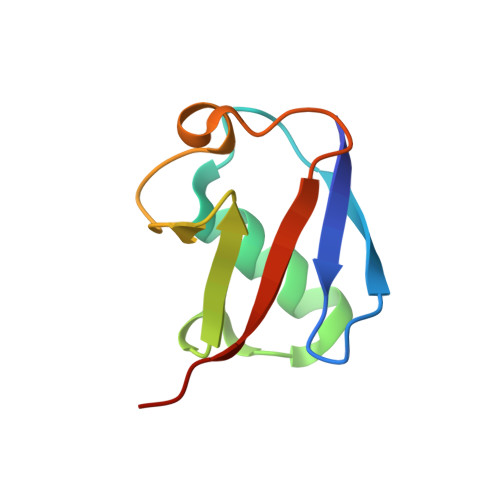
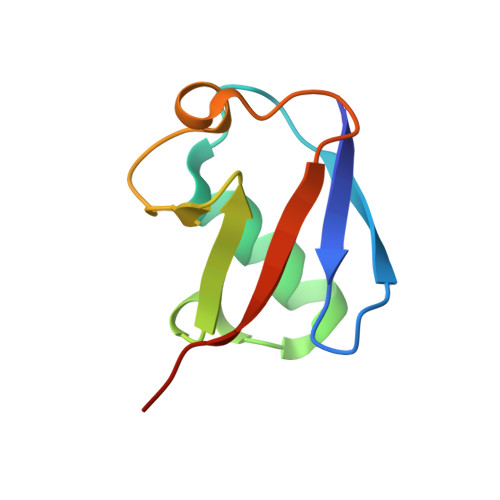
Structural basis of ubiquitin recognition by the winged-helix domain of Cockayne syndrome group B protein.
Takahashi, T.S., Sato, Y., Yamagata, A., Goto-Ito, S., Saijo, M., Fukai, S.(2019) Nucleic Acids Res 47: 3784-3794
- PubMed: 30753618
- DOI: https://doi.org/10.1093/nar/gkz081
- Primary Citation of Related Structures:
6A6I - PubMed Abstract:
Cockayne syndrome group B (CSB, also known as ERCC6) protein is involved in many DNA repair processes and essential for transcription-coupled repair (TCR). The central region of CSB has the helicase motif, whereas the C-terminal region contains important regulatory elements for repair of UV- and oxidative stress-induced damages and double-strand breaks (DSBs). A previous study suggested that a small part (∼30 residues) within this region was responsible for binding to ubiquitin (Ub). Here, we show that the Ub-binding of CSB requires a larger part of CSB, which was previously identified as a winged-helix domain (WHD) and is involved in the recruitment of CSB to DSBs. We also present the crystal structure of CSB WHD in complex with Ub. CSB WHD folds as a single globular domain, defining a class of Ub-binding domains (UBDs) different from 23 UBD classes identified so far. The second α-helix and C-terminal extremity of CSB WHD interact with Ub. Together with structure-guided mutational analysis, we identified the residues critical for the binding to Ub. CSB mutants defective in the Ub binding reduced repair of UV-induced damage. This study supports the notion that DSB repair and TCR may be associated with the Ub-binding of CSB.
- Institute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan.
Organizational Affiliation: