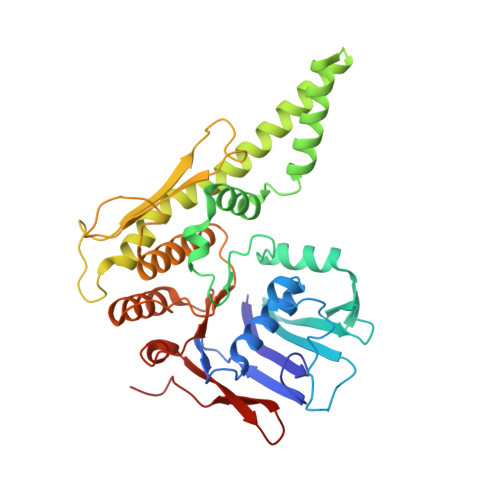
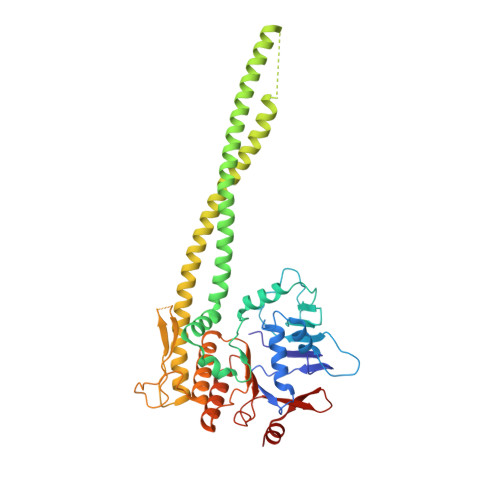
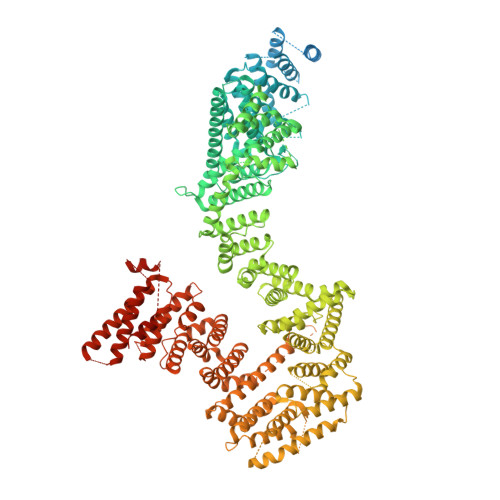
Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3.
Collier, J.E., Lee, B.G., Roig, M.B., Yatskevich, S., Petela, N.J., Metson, J., Voulgaris, M., Gonzalez Llamazares, A., Lowe, J., Nasmyth, K.A.(2020) Elife 9
- PubMed: 32930661
- DOI: https://doi.org/10.7554/eLife.59560
- Primary Citation of Related Structures:
6ZZ6 - PubMed Abstract:
In addition to extruding DNA loops, cohesin entraps within its SMC-kleisin ring (S-K) individual DNAs during G1 and sister DNAs during S-phase. All three activities require related hook-shaped proteins called Scc2 and Scc3. Using thiol-specific crosslinking we provide rigorous proof of entrapment activity in vitro. Scc2 alone promotes entrapment of DNAs in the E-S and E-K compartments, between ATP-bound engaged heads and the SMC hinge and associated kleisin, respectively. This does not require ATP hydrolysis nor is it accompanied by entrapment within S-K rings, which is a slower process requiring Scc3. Cryo-EM reveals that DNAs transported into E-S/E-K compartments are 'clamped' in a sub-compartment created by Scc2's association with engaged heads whose coiled coils are folded around their elbow. We suggest that clamping may be a recurrent feature of cohesin complexes active in loop extrusion and that this conformation precedes the S-K entrapment required for sister chromatid cohesion.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: