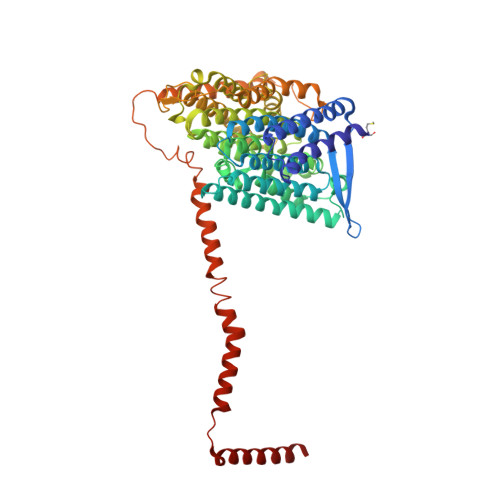
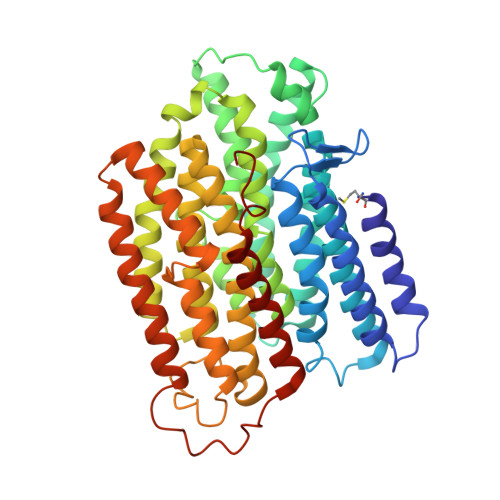
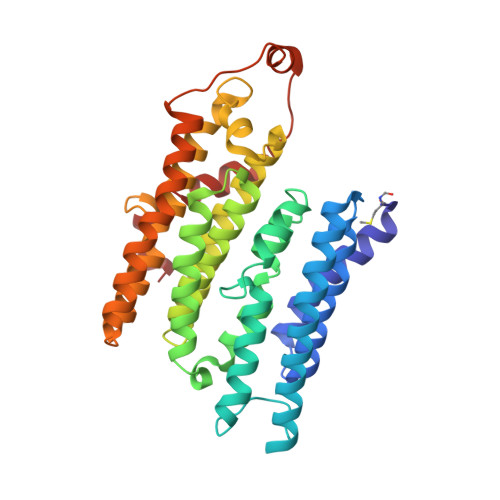
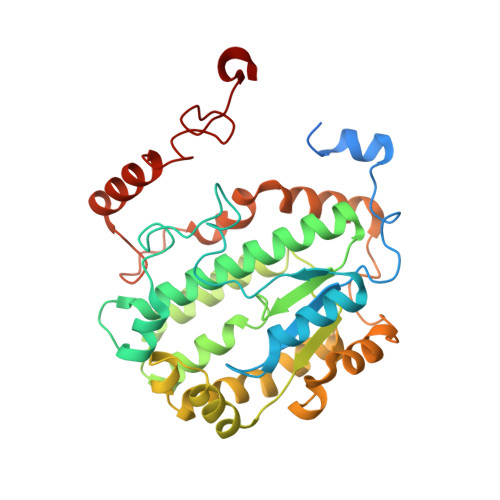
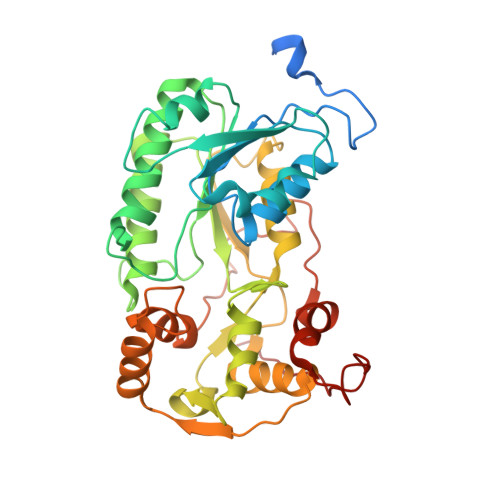
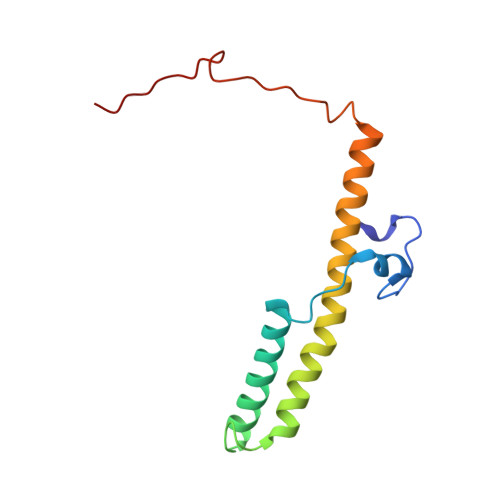
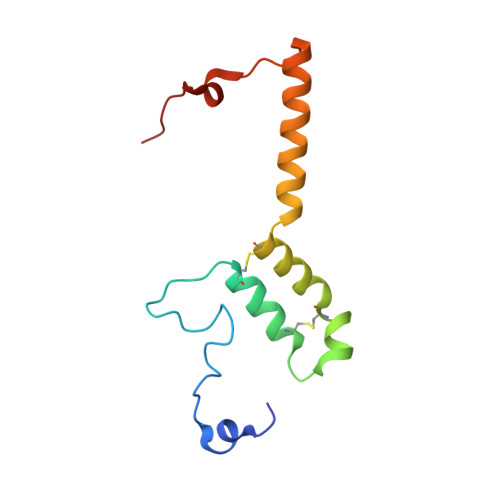
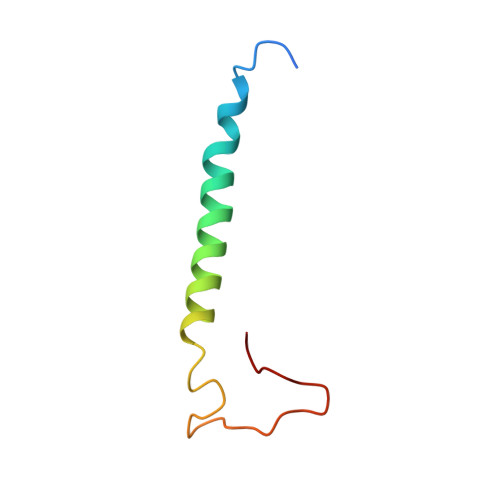
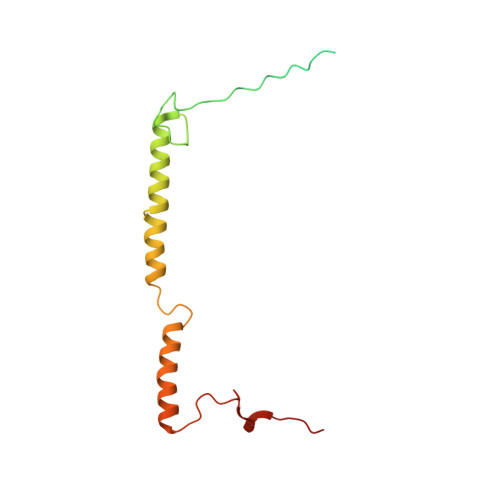
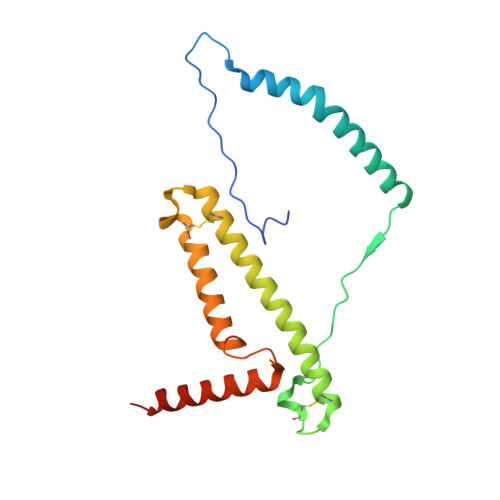
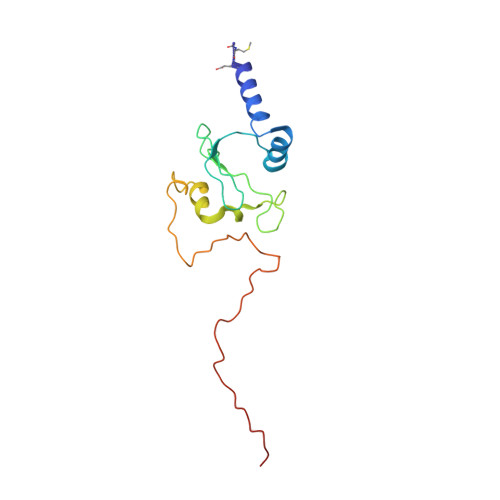
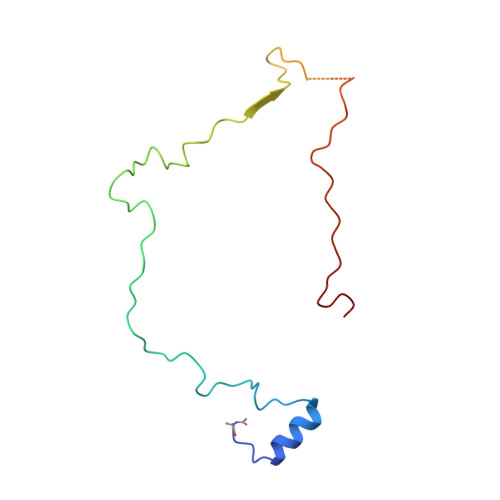
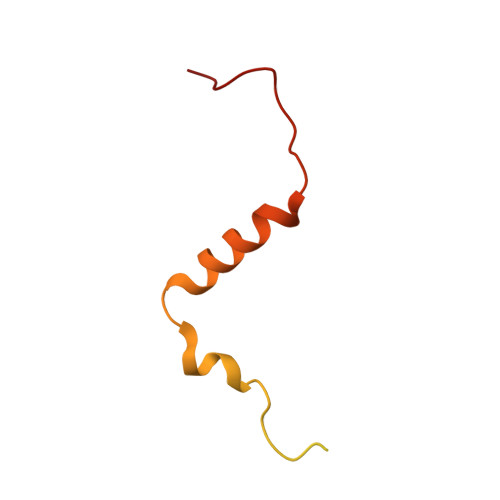
Structure of inhibitor-bound mammalian complex I.
Bridges, H.R., Fedor, J.G., Blaza, J.N., Di Luca, A., Jussupow, A., Jarman, O.D., Wright, J.J., Agip, A.A., Gamiz-Hernandez, A.P., Roessler, M.M., Kaila, V.R.I., Hirst, J.(2020) Nat Commun 11: 5261-5261
- PubMed: 33067417
- DOI: https://doi.org/10.1038/s41467-020-18950-3
- Primary Citation of Related Structures:
6ZR2, 6ZTQ - PubMed Abstract:
Respiratory complex I (NADH:ubiquinone oxidoreductase) captures the free energy from oxidising NADH and reducing ubiquinone to drive protons across the mitochondrial inner membrane and power oxidative phosphorylation. Recent cryo-EM analyses have produced near-complete models of the mammalian complex, but leave the molecular principles of its long-range energy coupling mechanism open to debate. Here, we describe the 3.0-Å resolution cryo-EM structure of complex I from mouse heart mitochondria with a substrate-like inhibitor, piericidin A, bound in the ubiquinone-binding active site. We combine our structural analyses with both functional and computational studies to demonstrate competitive inhibitor binding poses and provide evidence that two inhibitor molecules bind end-to-end in the long substrate binding channel. Our findings reveal information about the mechanisms of inhibition and substrate reduction that are central for understanding the principles of energy transduction in mammalian complex I.
- The Medical Research Council Mitochondrial Biology Unit, University of Cambridge, The Keith Peters Building, Cambridge Biomedical Campus, Hills Road, Cambridge, CB2 0XY, UK.
Organizational Affiliation: