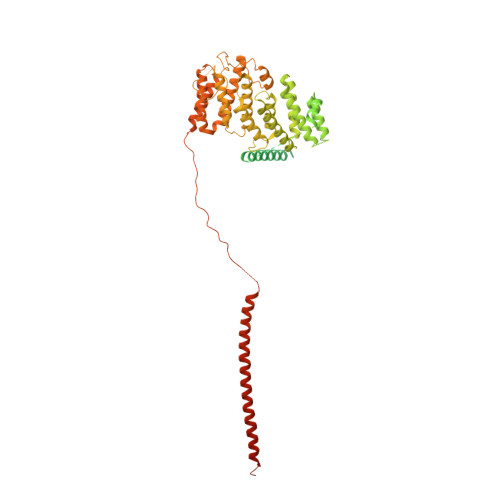
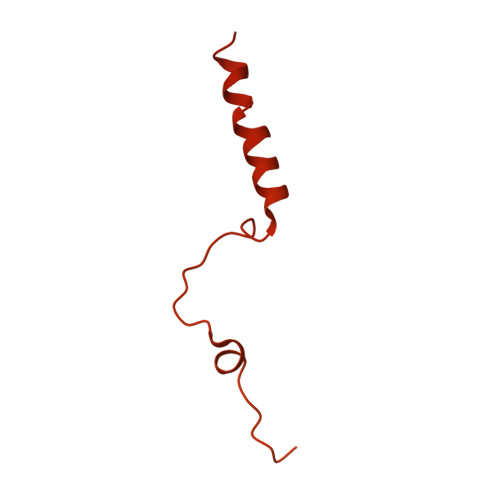
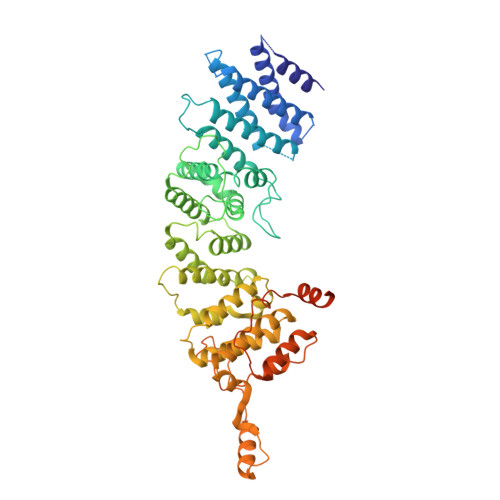
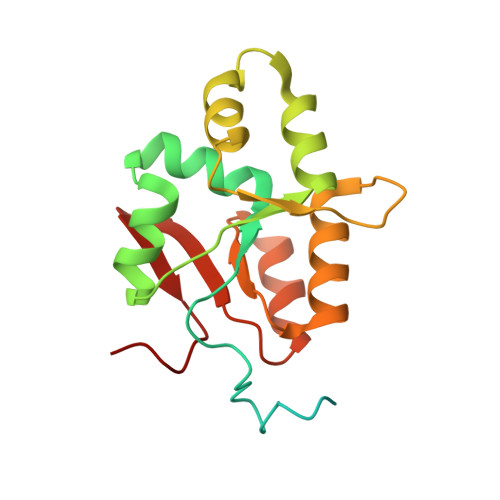
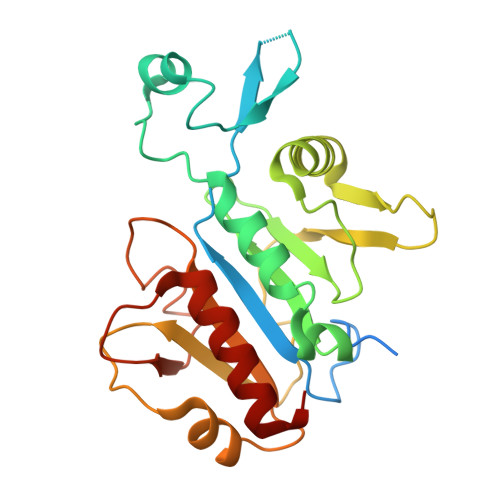
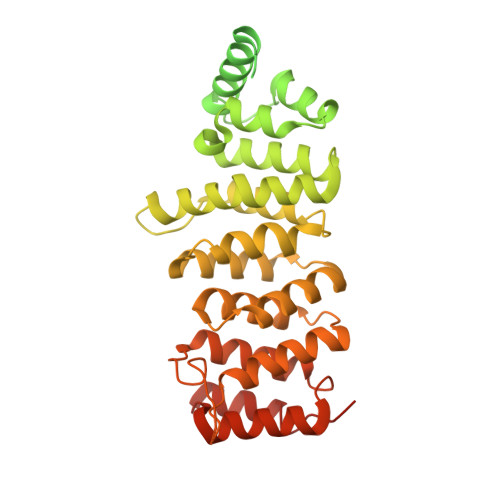
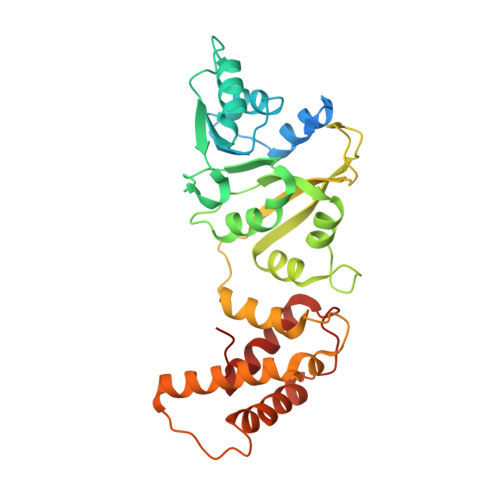
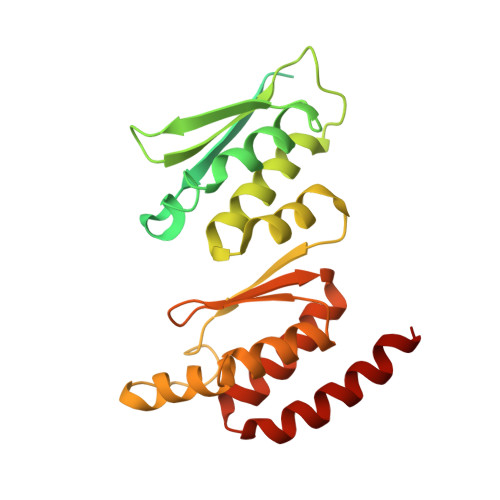
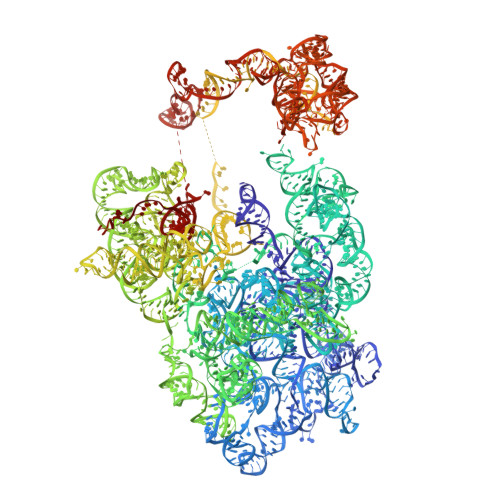
90 S pre-ribosome transformation into the primordial 40 S subunit.
Cheng, J., Lau, B., La Venuta, G., Ameismeier, M., Berninghausen, O., Hurt, E., Beckmann, R.(2020) Science 369: 1470-1476
- PubMed: 32943521
- DOI: https://doi.org/10.1126/science.abb4119
- Primary Citation of Related Structures:
6ZQA, 6ZQB, 6ZQC, 6ZQD, 6ZQE, 6ZQF, 6ZQG - PubMed Abstract:
Production of small ribosomal subunits initially requires the formation of a 90 S precursor followed by an enigmatic process of restructuring into the primordial pre-40 S subunit. We elucidate this process by biochemical and cryo-electron microscopy analysis of intermediates along this pathway in yeast. First, the remodeling RNA helicase Dhr1 engages the 90 S pre-ribosome, followed by Utp24 endonuclease-driven RNA cleavage at site A 1 , thereby separating the 5'-external transcribed spacer (ETS) from 18 S ribosomal RNA. Next, the 5'-ETS and 90 S assembly factors become dislodged, but this occurs sequentially, not en bloc. Eventually, the primordial pre-40 S emerges, still retaining some 90 S factors including Dhr1, now ready to unwind the final small nucleolar U3-18 S RNA hybrid. Our data shed light on the elusive 90 S to pre-40 S transition and clarify the principles of assembly and remodeling of large ribonucleoproteins.
- Gene Center, Department of Biochemistry, University of Munich, 81377 Munich, Germany.
Organizational Affiliation: