Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
Man, S., Redman, J.E., Cross, D.L., Cole, D.K., Can, I., Davies, B., Hashimdeen, S.S., Reid, R., Llewellyn-Lacey, S., Miners, K.L., Ladell, K., Lissina, A., Brown, P.E., Wooldridge, L., Price, D.A., Rizkallah, P.J.(2021) J Immunol 207: 1009-1017
- PubMed: 34321228
- DOI: https://doi.org/10.4049/jimmunol.2000756
- Primary Citation of Related Structures:
6Z9V, 6Z9W, 6Z9X - PubMed Abstract:
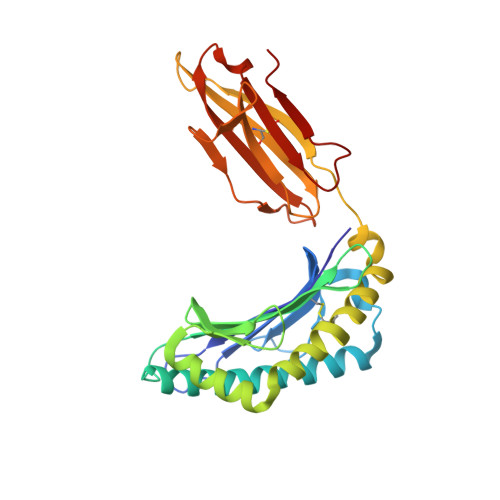
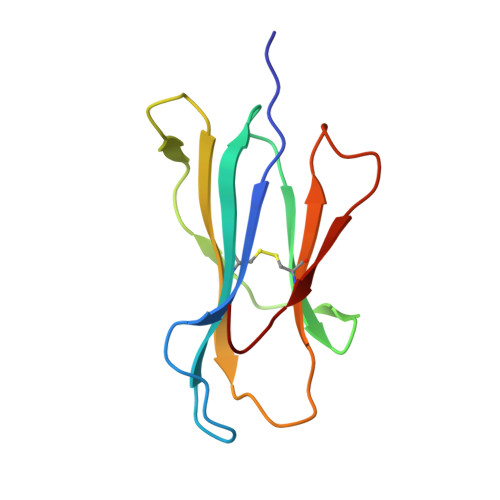
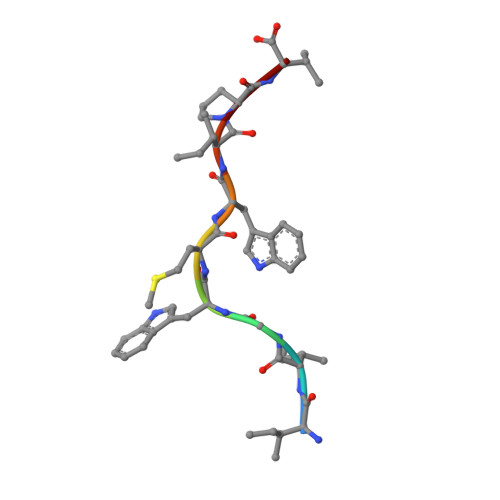
The human CD8 + T cell clone 6C5 has previously been shown to recognize the tert -butyl-modified Bax 161-170 peptide LLSY(3- t Bu)FGTPT presented by HLA-A*02:01. This nonnatural epitope was likely created as a by-product of fluorenylmethoxycarbonyl protecting group peptide synthesis and bound poorly to HLA-A*02:01. In this study, we used a systematic approach to identify and characterize natural ligands for the 6C5 TCR. Functional analyses revealed that 6C5 T cells only recognized the LLSYFGTPT peptide when t Bu was added to the tyrosine residue and did not recognize the LLSYFGTPT peptide modified with larger (di- t Bu) or smaller chemical groups (Me). Combinatorial peptide library screening further showed that 6C5 T cells recognized a series of self-derived peptides with dissimilar amino acid sequences to LLSY(3- t Bu)FGTPT. Structural studies of LLSY(3- t Bu)FGTPT and two other activating nonamers (IIGWMWIPV and LLGWVFAQV) in complex with HLA-A*02:01 demonstrated similar overall peptide conformations and highlighted the importance of the position (P) 4 residue for T cell recognition, particularly the capacity of the bulky amino acid tryptophan to substitute for the t Bu-modified tyrosine residue in conjunction with other changes at P5 and P6. Collectively, these results indicated that chemical modifications directly altered the immunogenicity of a synthetic peptide via molecular mimicry, leading to the inadvertent activation of a T cell clone with unexpected and potentially autoreactive specificities.
- Division of Cancer and Genetics, School of Medicine, Cardiff University, Cardiff, United Kingdom; ManS@cf.ac.uk.
Organizational Affiliation: