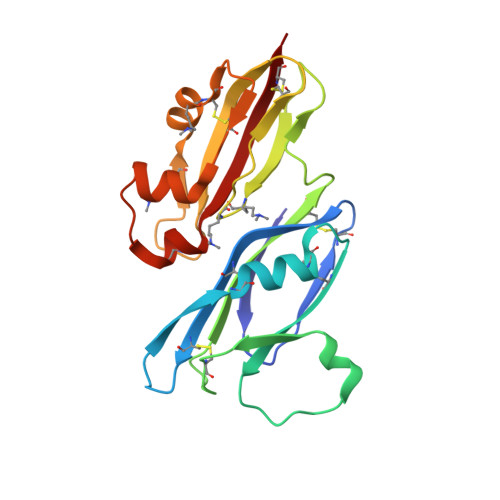
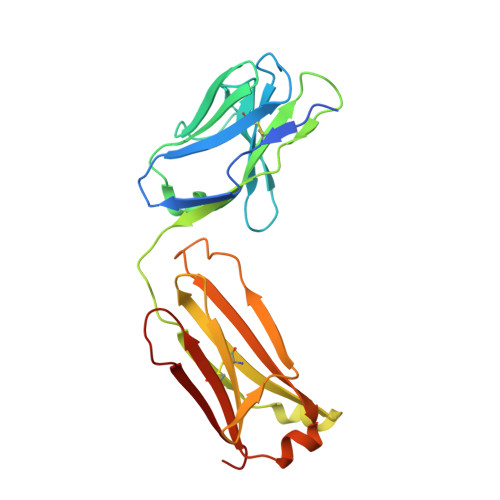
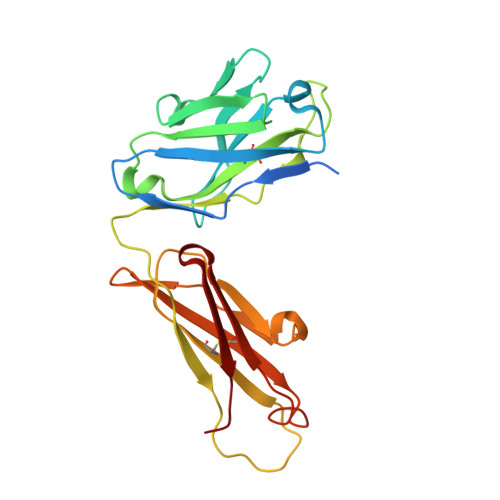
The Structure of the Cysteine-Rich Domain of Plasmodium falciparum P113 Identifies the Location of the RH5 Binding Site.
Campeotto, I., Galaway, F., Mehmood, S., Barfod, L.K., Quinkert, D., Kotraiah, V., Phares, T.W., Wright, K.E., Snijders, A.P., Draper, S.J., Higgins, M.K., Wright, G.J.(2020) mBio 11
- PubMed: 32900802
- DOI: https://doi.org/10.1128/mBio.01566-20
- Primary Citation of Related Structures:
6Z2L - PubMed Abstract:
Plasmodium falciparum RH5 is a secreted parasite ligand that is essential for erythrocyte invasion through direct interaction with the host erythrocyte receptor basigin. RH5 forms a tripartite complex with two other secreted parasite proteins, CyRPA and RIPR, and is tethered to the surface of the parasite through membrane-anchored P113. Antibodies against RH5, CyRPA, and RIPR can inhibit parasite invasion, suggesting that vaccines containing these three components have the potential to prevent blood-stage malaria. To further explore the role of the P113-RH5 interaction, we selected monoclonal antibodies against P113 that were either inhibitory or noninhibitory for RH5 binding. Using a Fab fragment as a crystallization chaperone, we determined the crystal structure of the RH5 binding region of P113 and showed that it is composed of two domains with structural similarities to rhamnose-binding lectins. We identified the RH5 binding site on P113 by using a combination of hydrogen-deuterium exchange mass spectrometry and site-directed mutagenesis. We found that a monoclonal antibody to P113 that bound to this interface and inhibited the RH5-P113 interaction did not inhibit parasite blood-stage growth. These findings provide further structural information on the protein interactions of RH5 and will be helpful in guiding the development of blood-stage malaria vaccines that target RH5. IMPORTANCE Malaria is a deadly infectious disease primarily caused by the parasite Plasmodium falciparum It remains a major global health problem, and there is no highly effective vaccine. A parasite protein called RH5 is centrally involved in the invasion of host red blood cells, making it-and the other parasite proteins it interacts with-promising vaccine targets. We recently identified a protein called P113 that binds RH5, suggesting that it anchors RH5 to the parasite surface. In this paper, we use structural biology to locate and characterize the RH5 binding region on P113. These findings will be important to guide the development of new antimalarial vaccines to ultimately prevent this disease, which affects some of the poorest people on the planet.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: