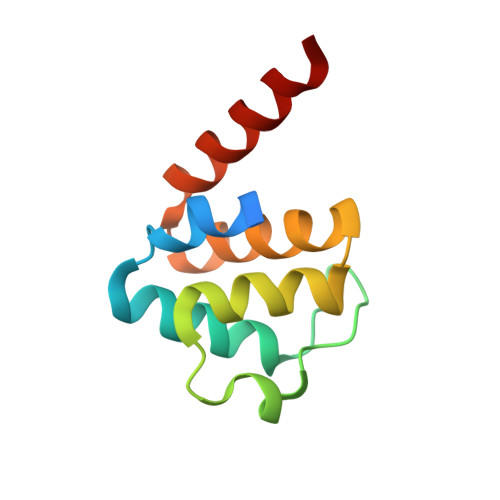
Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly.
Marleaux, M., Anand, K., Latz, E., Geyer, M.(2020) FEBS Lett 594: 2383-2395
- PubMed: 32542665
- DOI: https://doi.org/10.1002/1873-3468.13865
- Primary Citation of Related Structures:
6Z2G - PubMed Abstract:
Inflammasomes are cytosolic multimeric signaling complexes of the innate immune system that induce activation of caspases. The NOD-like receptor NLRP9 recruits the adaptor protein ASC to form an ASC-dependent inflammasome to limit rotaviral replication in intestinal epithelial cells, but only little is known about the molecular mechanisms regulating and driving its assembly. Here, we present the crystal structure of the human NLRP9 pyrin domain (PYD). We show that NLRP9 PYD is not able to self-polymerize nor to nucleate ASC specks in HEK293T cells. A comparison with filament-forming PYDs revealed that NLRP9 PYD adopts a conformation compatible with filament formation, but several charge inversions of interfacing residues might cause repulsive effects that prohibit self-oligomerization. These results propose that inflammasome assembly of NLRP9 might differ largely from what we know of other inflammasomes.
- Institute of Structural Biology, University of Bonn, Bonn, Germany.
Organizational Affiliation: