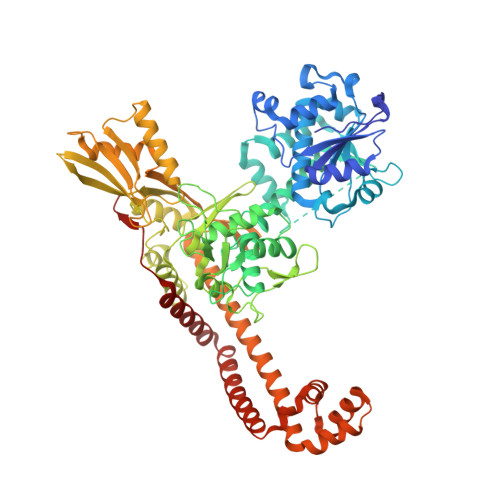
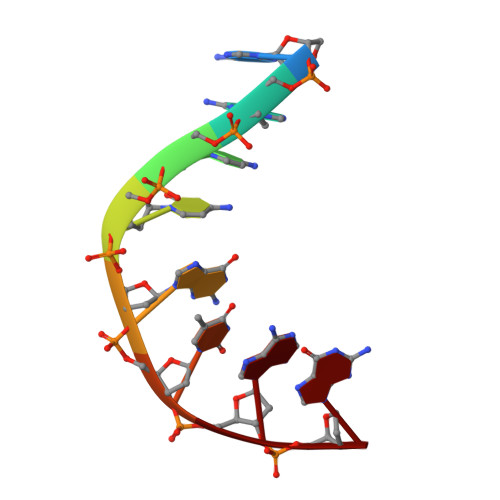
Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds.
Kolaric, A., Germe, T., Hrast, M., Stevenson, C.E.M., Lawson, D.M., Burton, N.P., Voros, J., Maxwell, A., Minovski, N., Anderluh, M.(2021) Nat Commun 12: 150-150
- PubMed: 33420011
- DOI: https://doi.org/10.1038/s41467-020-20405-8
- Primary Citation of Related Structures:
6Z1A - PubMed Abstract:
Novel bacterial type II topoisomerase inhibitors (NBTIs) stabilize single-strand DNA cleavage breaks by DNA gyrase but their exact mechanism of action has remained hypothetical until now. We have designed a small library of NBTIs with an improved DNA gyrase-binding moiety resulting in low nanomolar inhibition and very potent antibacterial activity. They stabilize single-stranded cleavage complexes and, importantly, we have obtained the crystal structure where an NBTI binds gyrase-DNA in a single conformation lacking apparent static disorder. This directly proves the previously postulated NBTI mechanism of action and shows that they stabilize single-strand cleavage through asymmetric intercalation with a shift of the scissile phosphate. This crystal stucture shows that the chlorine forms a halogen bond with the backbone carbonyls of the two symmetry-related Ala68 residues. To the best of our knowledge, such a so-called symmetrical bifurcated halogen bond has not been identified in a biological system until now.
- Theory Department, Laboratory for Cheminformatics, National Institute of Chemistry, Hajdrihova 19, 1001, Ljubljana, Slovenia.
Organizational Affiliation: