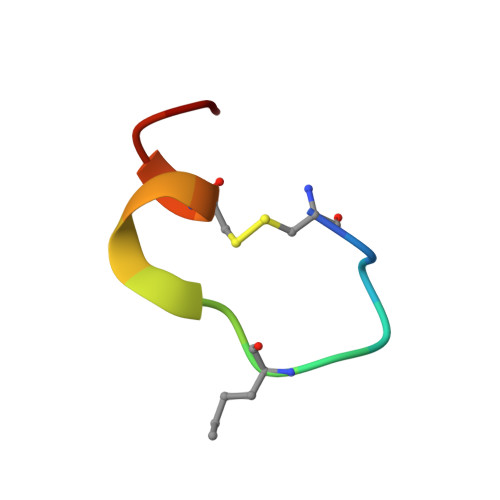
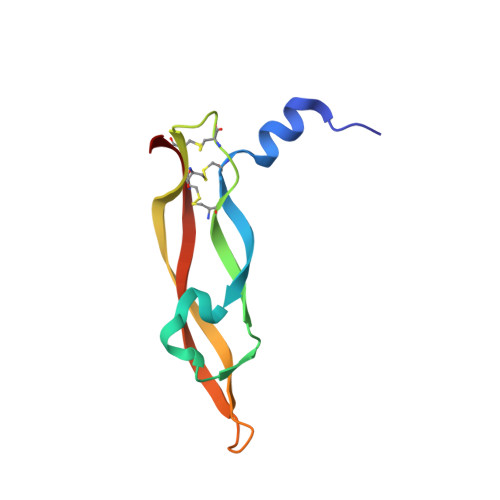
Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Gaucher, J.F., Reille-Seroussi, M., Broussy, S.(2022) Chemistry
- PubMed: 35665969
- DOI: https://doi.org/10.1002/chem.202200465
- Primary Citation of Related Structures:
6Z13, 6Z3F, 6ZBR, 6ZCD, 6ZFL - PubMed Abstract:
Macrocyclization constraints are widely used in the design of protein ligands to stabilize their bioactive conformation and increase their affinities. However, the resulting changes in binding entropy can be puzzling and uncorrelated to affinity gains. Here, the thermodynamic (Isothermal Titration Calorimetry) and structural (X-ray, NMR and CD) analysis of a complete series of lactam-bridged peptide ligands of the vascular endothelial growth factor, and their unconstrained analogs are reported. It is shown that differences in thermodynamics arise mainly from the folding energy of the peptide upon binding. The systematic reduction in conformational entropy penalty due to helix pre-organization can be counterbalanced by an unfavorable vibrational entropy change if the constraints are too rigid. The gain in configurational entropy partially escapes the enthalpy/entropy compensation and leads to an improvement in affinity. The precision of the analytical ITC method makes this study a possible benchmark for constrained peptides optimization.
- CiTCoM, UMR CNRS 8038, Université Paris Cité, Faculté de Santé, UFR de Pharmacie, 4 av. de l'Observatoire, 75006, Paris, France.
Organizational Affiliation: