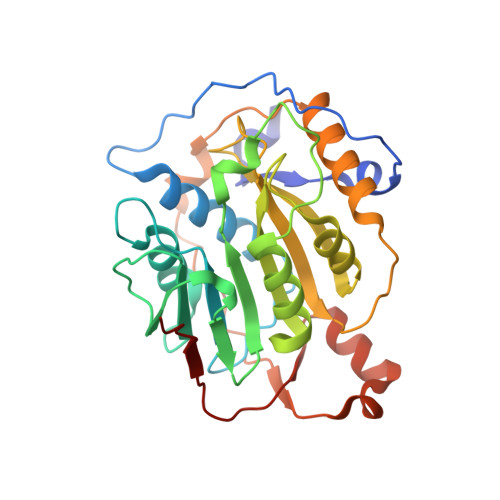
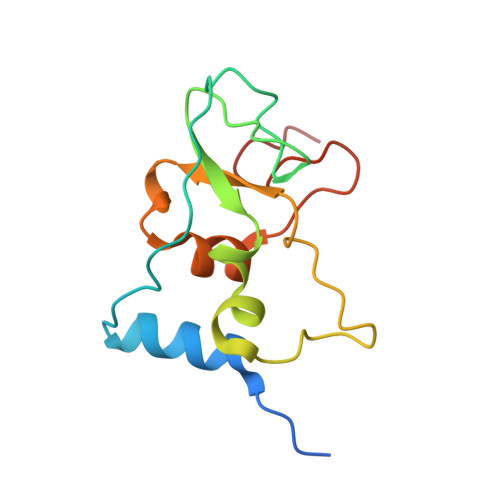
Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin.
Krafcikova, P., Silhan, J., Nencka, R., Boura, E.(2020) Nat Commun 11: 3717-3717
- PubMed: 32709887
- DOI: https://doi.org/10.1038/s41467-020-17495-9
- Primary Citation of Related Structures:
6YZ1 - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the cause of the COVID-19 pandemic. 2'-O-RNA methyltransferase (MTase) is one of the enzymes of this virus that is a potential target for antiviral therapy as it is crucial for RNA cap formation; an essential process for viral RNA stability. This MTase function is associated with the nsp16 protein, which requires a cofactor, nsp10, for its proper activity. Here we show the crystal structure of the nsp10-nsp16 complex bound to the pan-MTase inhibitor sinefungin in the active site. Our structural comparisons reveal low conservation of the MTase catalytic site between Zika and SARS-CoV-2 viruses, but high conservation of the MTase active site between SARS-CoV-2 and SARS-CoV viruses; these data suggest that the preparation of MTase inhibitors targeting several coronaviruses - but not flaviviruses - should be feasible. Together, our data add to important information for structure-based drug discovery.
- Institute of Organic Chemistry and Biochemistry AS CR, v.v.i., Flemingovo nam. 2., 166 10, Prague 6, Czech Republic.
Organizational Affiliation: