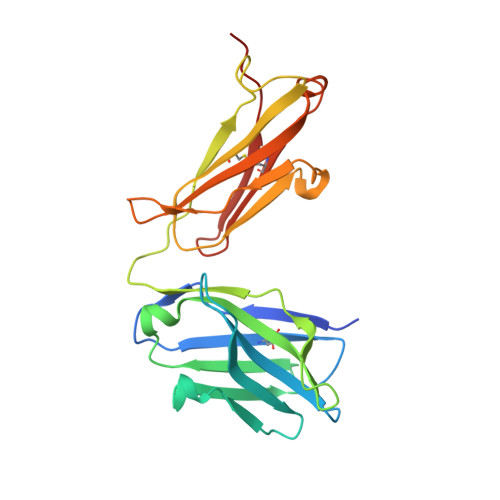
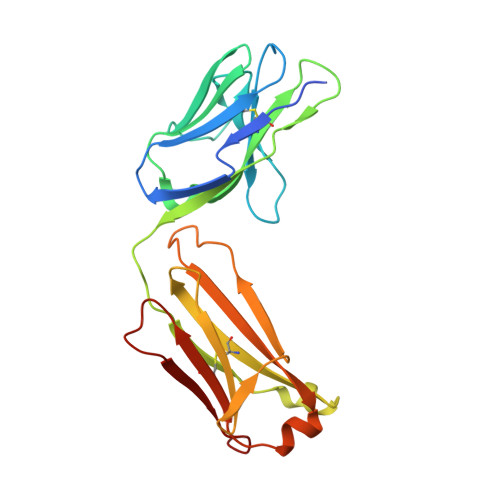
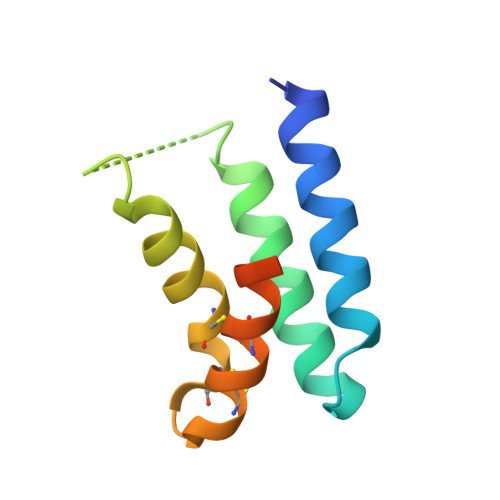
Bottom-up de novo design of functional proteins with complex structural features.
Yang, C., Sesterhenn, F., Bonet, J., van Aalen, E.A., Scheller, L., Abriata, L.A., Cramer, J.T., Wen, X., Rosset, S., Georgeon, S., Jardetzky, T., Krey, T., Fussenegger, M., Merkx, M., Correia, B.E.(2021) Nat Chem Biol 17: 492-500
- PubMed: 33398169
- DOI: https://doi.org/10.1038/s41589-020-00699-x
- Primary Citation of Related Structures:
6YWC, 6YWD - PubMed Abstract:
De novo protein design has enabled the creation of new protein structures. However, the design of functional proteins has proved challenging, in part due to the difficulty of transplanting structurally complex functional sites to available protein structures. Here, we used a bottom-up approach to build de novo proteins tailored to accommodate structurally complex functional motifs. We applied the bottom-up strategy to successfully design five folds for four distinct binding motifs, including a bifunctionalized protein with two motifs. Crystal structures confirmed the atomic-level accuracy of the computational designs. These de novo proteins were functional as components of biosensors to monitor antibody responses and as orthogonal ligands to modulate synthetic signaling receptors in engineered mammalian cells. Our work demonstrates the potential of bottom-up approaches to accommodate complex structural motifs, which will be essential to endow de novo proteins with elaborate biochemical functions, such as molecular recognition or catalysis.
- Institute of Bioengineering, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland.
Organizational Affiliation: