Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity.
Shin, D., Mukherjee, R., Grewe, D., Bojkova, D., Baek, K., Bhattacharya, A., Schulz, L., Widera, M., Mehdipour, A.R., Tascher, G., Geurink, P.P., Wilhelm, A., van der Heden van Noort, G.J., Ovaa, H., Muller, S., Knobeloch, K.P., Rajalingam, K., Schulman, B.A., Cinatl, J., Hummer, G., Ciesek, S., Dikic, I.(2020) Nature 587: 657-662
- PubMed: 32726803
- DOI: https://doi.org/10.1038/s41586-020-2601-5
- Primary Citation of Related Structures:
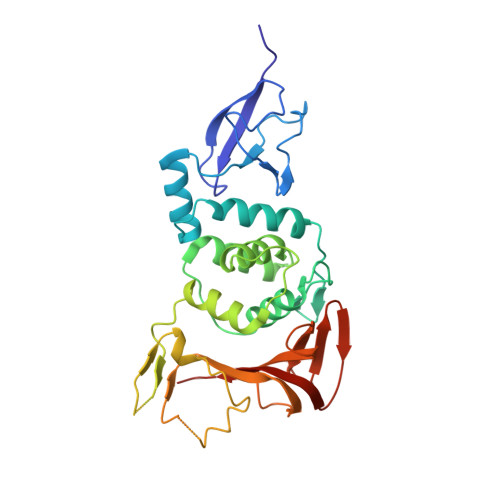
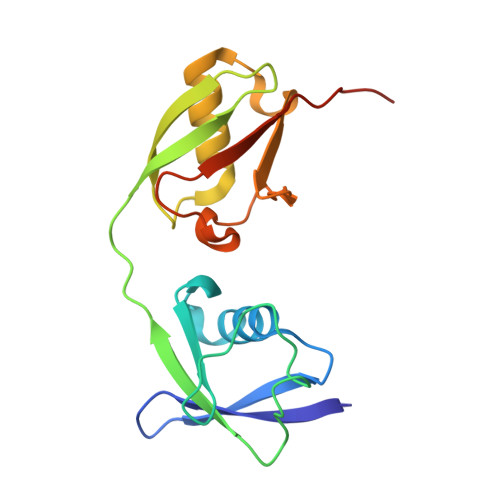
6YVA - PubMed Abstract:
The papain-like protease PLpro is an essential coronavirus enzyme that is required for processing viral polyproteins to generate a functional replicase complex and enable viral spread 1,2 . PLpro is also implicated in cleaving proteinaceous post-translational modifications on host proteins as an evasion mechanism against host antiviral immune responses 3-5 . Here we perform biochemical, structural and functional characterization of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) PLpro (SCoV2-PLpro) and outline differences with SARS-CoV PLpro (SCoV-PLpro) in regulation of host interferon and NF-κB pathways. SCoV2-PLpro and SCoV-PLpro share 83% sequence identity but exhibit different host substrate preferences; SCoV2-PLpro preferentially cleaves the ubiquitin-like interferon-stimulated gene 15 protein (ISG15), whereas SCoV-PLpro predominantly targets ubiquitin chains. The crystal structure of SCoV2-PLpro in complex with ISG15 reveals distinctive interactions with the amino-terminal ubiquitin-like domain of ISG15, highlighting the high affinity and specificity of these interactions. Furthermore, upon infection, SCoV2-PLpro contributes to the cleavage of ISG15 from interferon responsive factor 3 (IRF3) and attenuates type I interferon responses. Notably, inhibition of SCoV2-PLpro with GRL-0617 impairs the virus-induced cytopathogenic effect, maintains the antiviral interferon pathway and reduces viral replication in infected cells. These results highlight a potential dual therapeutic strategy in which targeting of SCoV2-PLpro can suppress SARS-CoV-2 infection and promote antiviral immunity.
- Institute of Biochemistry II, Faculty of Medicine, Goethe University, Frankfurt, Germany.
Organizational Affiliation: