Crystal structure of the Diels Alderase AbmU from Streptomyces koyangensis
Back, C.R., Mbatha, S., Johns, S., Burton, N., Tiwari, K., van der Kamp, M., Willis, C., Race, P.R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AbmU | A [auth AAA], B [auth BBB], C [auth CCC], D [auth DDD] | 237 | Streptomyces koyangensis | Mutation(s): 0 | 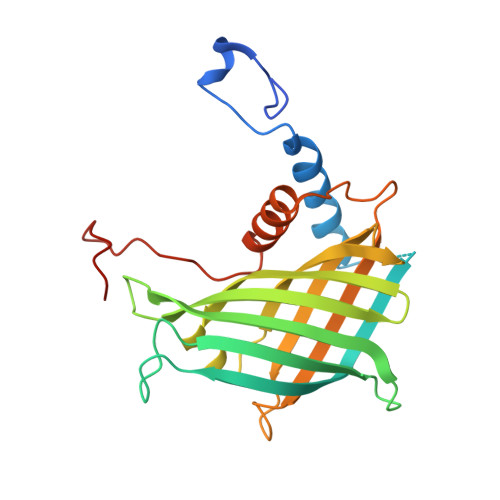 |
UniProt | |||||
Find proteins for A0A2P1BT29 (Streptomyces koyangensis) Explore A0A2P1BT29 Go to UniProtKB: A0A2P1BT29 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2P1BT29 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BR Query on BR | E [auth AAA] F [auth AAA] G [auth BBB] H [auth BBB] I [auth CCC] | BROMIDE ION Br CPELXLSAUQHCOX-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 66.128 | α = 90 |
| b = 64.998 | β = 93.813 |
| c = 97.865 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DIALS | data reduction |
| xia2 | data scaling |
| CRANK2 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/T001968/1 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/M012107/1 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/L01386X/1 |
| Engineering and Physical Sciences Research Council | United Kingdom | EP/G0367641/1 |